doi: 10.56294/saludcyt20241079
ORIGINAL
Brain tumour detection via EfficientDet and classification with DynaQ-GNN-LSTM
Detección de tumores cerebrales mediante EfficientDet y clasificación con DynaQ-GNN-LSTM
Ayesha Agrawal1 ![]() *, Vinod Maan1
*, Vinod Maan1 ![]() *
*
1Mody University of Science & Technology, Computer Science & Engineering. Lakshmangarh, India.
Cite as: Agrawal A, Maan V. Brain tumour detection via EfficientDet and classification with DynaQ-GNN-LSTM Salud, Ciencia y Tecnología. 2024; 4:1079. https://doi.org/10.56294/saludcyt20241079
Submitted: 07-02-2024 Revised: 25-04-2024 Accepted: 30-06-2024 Published: 01-07-2024
Editor:
Dr.
William Castillo-González ![]()
ABSTRACT
The early detection and accurate staging of brain tumors are critical for effective treatment strategies and improving patient outcomes. Existing methods for brain tumor classification often struggle with limitations such as suboptimal precision, accuracy, and recall rates, alongside significant delays in processing. The current methodologies in brain tumor classification frequently encounter issues such as inadequate feature extraction capabilities and limited accuracy in segmentation, which impede their effectiveness. To address these challenges, the proposed model integrates Fuzzy C-Means for segmentation, leveraging its ability to enhance the accuracy in distinguishing tumor regions. Bounding boxes surrounding identified tumour regions are produced by the method by efficiently utilising calculated region attributes. The use of Vision Transformers for feature extraction marks a significant advancement, offering a more nuanced analysis of the intricate patterns within brain imaging data samples. These features are then classified using a Dyna Q Graph LSTM (DynaQ-GNN-LSTM), a cutting-edge approach that combines the strengths of deep learning, reinforcement learning, and graph neural networks. The superiority of the proposed model is evident through its performance on multiple datasets. It demonstrates an 8,3 % increase in precision, 8,5 % increase in accuracy, 4,9 % increase in recall and 4,5 % increase in specificity, alongside 2,9 % reduction in delay compared to existing methods. In conclusion, the proposed method offers an efficient solution to the challenges faced in brain tumor classification. The study’s findings underscore the transformative impact of integrating cutting-edge technologies in medical diagnostics, paving the way for more accurate, and timely health interventions for clinical scenarios.
Keywords: Brain Tumor Classification; Fuzzy C-Means; Vision Transformers; Dyna-Q learning; Brain Tumor Detection; EfficientDet.
RESUMEN
La detección precoz y la estadificación precisa de los tumores cerebrales son fundamentales para aplicar estrategias de tratamiento eficaces y mejorar la evolución de los pacientes. Los métodos existentes para la clasificación de tumores cerebrales a menudo se enfrentan a limitaciones tales como tasas de precisión, exactitud y recuperación subóptimas, junto con retrasos significativos en el procesamiento. Las metodologías actuales de clasificación de tumores cerebrales se enfrentan con frecuencia a problemas como una capacidad inadecuada de extracción de características y una precisión limitada en la segmentación, que impiden su eficacia. Para hacer frente a estos problemas, el modelo propuesto integra Fuzzy C-Means para la segmentación, aprovechando su capacidad para mejorar la precisión en la distinción de las regiones tumorales. El método produce cuadros delimitadores alrededor de las regiones tumorales identificadas utilizando eficazmente los atributos de región calculados. El uso de transformadores de visión para la extracción de características supone un avance significativo, ya que ofrece un análisis más matizado de los intrincados patrones de las muestras de datos de imágenes cerebrales. A continuación, estas características se clasifican utilizando una Dyna Q Graph LSTM (DynaQ-GNN-LSTM), un enfoque de vanguardia que combina los puntos fuertes del aprendizaje profundo, el aprendizaje por refuerzo y las redes neuronales gráficas. La superioridad del modelo propuesto es evidente a través de su rendimiento en múltiples conjuntos de datos. Demuestra un aumento del 8,3 % en la precisión, un aumento del 8,5 % en la exactitud, un aumento del 4,9 % en el recuerdo y un aumento del 4,5 % en la especificidad, junto con una reducción del 2,9 % en el retraso en comparación con los métodos existentes. En conclusión, el método propuesto ofrece una solución eficaz a los retos que plantea la clasificación de tumores cerebrales. Los resultados del estudio subrayan el impacto transformador de la integración de tecnologías de vanguardia en el diagnóstico médico, allanando el camino para intervenciones sanitarias más precisas y oportunas para escenarios clínicos.
Palabras clave: Clasificación de Tumores Cerebrales; Fuzzy C-Means; Transformadores de Visión; Aprendizaje Dyna-Q; Detección de Tumores Cerebrales; EfficientDet.
INTRODUCTION
The realm of medical imaging and diagnostics has experienced significant advancements in recent years, particularly in the area of brain tumor identification and classification. Accurate and early detection of brain tumors is paramount for effective treatment planning and improving patient survival rates. However, the complexity of brain structures and the subtle variations in tumor appearances pose considerable challenges in accurate tumor classification and staging. This paper introduces a novel computational framework designed to address these challenges, offering significant improvements in precision, accuracy, recall, and processing speed.
Brain tumors are characterized by abnormal growths of cells within or around the brain and can vary widely in size, location, and malignancy. The traditional methods of brain tumor classification, primarily reliant on manual interpretation of Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) scans by radiologists, are time-consuming and subject to human error. Automated classification systems have been developed to assist in this process, but they often struggle with issues like low specificity, high false-positive rates, and limited ability to distinguish between tumor stages.
The introduction of machine learning and deep learning techniques has revolutionized the field of medical image analysis. These methods offer the potential for more accurate and efficient analysis by automatically learning complex patterns within the imaging data samples. However, existing models still face limitations in terms of feature extraction and segmentation accuracy, which are crucial for precise tumor classification. Furthermore, the dynamic and heterogeneous nature of brain tumor imaging data necessitates a model that can adapt and learn from diverse and complex data structures.
In response to these challenges, our research utilizes EfficientDet for brain tumor detection. When compared to other techniques, EfficientDet maintains computing efficiency while achieving higher levels of accuracy. Through the use of effective design decisions like the BiFPN and efficient head architecture, it is able to attain improved performance without appreciably raising computational costs. Additionally for classification purpose, our research proposes a comprehensive approach that synergizes Fuzzy C-Means (FCM) segmentation, Vision Transformers for feature extraction, and a Dyna Q Graph LSTM(DYNAQ-GNN-LSTM ) for feature classification. FCM is employed for its efficacy in enhancing image segmentation, particularly in delineating tumor regions with high precision. Vision Transformers are utilized for feature extraction, capitalizing on their ability to capture detailed and comprehensive patterns in imaging data samples. The final classification is executed by the DYNAQ-GNN-LSTM, which combines the strengths of deep learning, reinforcement learning, and graph neural networks, facilitating a more dynamic and context-aware understanding of the imaging data samples.
The detection technique attained Recall of 98,95 % an accuracy of 99,50 % along with Specificity of 98,99 % and precision of 99 %. Our classification approach has been rigorously tested on multiple datasets, displaying remarkable improvements over existing methods. The results demonstrate an 8,3 % increase in precision, an 8,5 % increase in accuracy, a 4,9 % increase in recall, a 3,9 % increase in the Area under the Curve (AUC), and a 4,5 % increase in specificity, along with a 2,9 % reduction in delay.
These advancements not only represent a significant leap in the technical capabilities of brain tumor classification systems but also hold profound clinical implications. This paper details the design and implementation of this innovative computational framework and discusses its implications in the broader context of medical imaging and diagnostics. The integration of cutting-edge technologies in this framework sets a new benchmark in the field, illustrating the transformative potential of advanced computational techniques in improving healthcare outcomes.
In summary, the motivation behind this research is to address the critical challenges in brain tumor diagnosis and to offer a solution that is not only technically superior but also has a meaningful impact on patient care. The contributions of this study are substantial, offering advancements in segmentation, feature extraction, and classification techniques, and demonstrating significant improvements in diagnostic accuracy and efficiency. This research represents a significant step forward in the application of advanced computational methods in medical imaging and neuro-oncology.
Literature review
The recent advancements in brain tumor classification and segmentation using various deep learning and neural network approaches have been substantial, as evidenced by a series of studies conducted in this field. Karaaltun’s work(1) demonstrates the efficacy of using a whole image average pooling-based convolutional neural network (CNN) for brain tumor classification. This approach highlights the potential for CNNs in analyzing medical images, particularly for complex tasks like tumor classification.
Parallelly, Choudhuri et al.(2) introduced a novel quantum classical CNN architecture for brain MRI tumor classification. This innovative approach signifies a leap in combining quantum computing principles with classical neural networks, aiming to elevate the accuracy and efficiency of classification. Similarly, Dixit et al.(3) explored the use of a novel statistical feature extractor in conjunction with relevance vector machines (RVM) for MRI image brain tumor classification, providing an alternative to traditional CNN methods.
Singh et al.(4) further corroborates the effectiveness of enhanced CNNs in automated brain tumor classification in MR images. Their work emphasizes the continual improvement and customization of CNN architectures for specific medical imaging tasks. In a similar vein, Balamurugan(5) proposed a hybrid deep CNN combined with a LuNetClassifier for brain tumor segmentation and classification, showcasing the trend towards hybrid deep learning models in medical imaging.
Expanding the scope of neural networks in this domain, Srividya et al.(6) introduced a Histo-Quartic Graph and Stack Entropy-Based Deep Neural Network method for brain and tumor segmentation. This approach illustrates the growing interest in combining graph-based methods with deep learning for enhanced image analysis. Karri et al.(7) presented the SGC-ARANet, a scale-wise global contextual axile reverse attention network, for automatic brain tumor segmentation, highlighting the use of attention mechanisms in neural networks for medical image analysis.
Islam et al.(8) examined the effectiveness of federated learning and CNN ensemble architectures in identifying brain tumors using MRI images. Their work points to the potential of distributed learning models and ensemble methods in improving diagnostic accuracy. Halder et al.(9) introduced a novel histogram feature for brain tumor detection, emphasizing the importance of feature engineering in enhancing the performance of machine learning models.
Alyami et al.(10) explored tumor localization and classification from MRI of the brain using a deep convolutional neural network and Salp Swarm Algorithm, indicating the integration of swarm intelligence with deep learning for medical imaging tasks. Yildirim et al.(11) focused on the detection and classification of various types of tumors present in the brain using a deep learning-based hybrid model, showcasing the versatility of deep learning models in distinguishing between different tumor types.
Saurav et al.(12) presented an attention-guided CNN for automated classification of brain tumors from MRI, further underscoring the utility of attention mechanisms in neural networks for medical imaging. In order to overcome the shortcomings of existing models, this research(13) presents a unique Graph Convolutional Neural Network (GCNN) model for brain tumour diagnosis and classification. Using a 26-layer CNN in conjunction with GNN, the method yields 95,01 % efficiency, with Net-2 surpassing the rest. This novel method presents a viable way to enhance the diagnostic and therapeutic management of brain tumours.
Asiri et al.(14) uses five pre-trained vision transformer models to examine the classification of images related to brain tumours. With an exceptional performance rate of 98,24 %, ViT-b32 outperforms various alternatives and current techniques. These findings highlight the usefulness of ViT models for medical image interpretation and establish a standard for further studies on brain tumour classification. In order to improve computer-assisted brain tumour diagnosis from MRI images, presents an improved Block-Wise Visual Geometry Group-19 (BW-VGG19) structure that achieves an accuracy rating of 98 %. This approach outperforms the current CNN and VGG16/19 approaches, answering the essential demand for precise and effective tumour diagnosis.(15)
M. Aggarwal et al.(16) addresses the difficulties caused by the intricate geometries of tumours in MRI scans by presenting an effective technique for brain tumour segmentation utilising an enhanced residual network (ResNet). In comparison to conventional CNN and FCN approaches, the methodology delivers superior results of more than ten percent enhancements in accuracy on MRI data, greater precision, and faster learning. This is achieved by optimising ResNet’s exchange of information and bypass connection. Presents Brain MRNet, a network using deep learning that can identify pituitary tumours, gliomas, and meningiomas with remarkable precision from MRI imaging. This strategy, which combines residual blocks and attention modules with an approach to segmentation for accurate tumour location, holds potential for assisting medical decisions in the identification of brain cancer.(17)
Uses YOLOv5’s object recognition skills to investigate brain tumour segmentation. The model demonstrates its potential for automated brain tumour detection by achieving comparable precision with several tumour classes: Glioma, Meningioma and Pituitary. This precision is critical for timely identification utilising MRI data.(18) Investigates the implementation of the EfficientDet model to real-time investigation of infrared footage from cameras with the goal of detecting autos, citizens, and deer near a classified site. Although there are obstacles related to real-time performance and limited data, the research suggests that this approach has the capacity to improve systems for monitoring.(19)
Abdusalomov et al.(20) used a sizable collection of brain tumour images to tackle the difficult problem of brain tumour detection in MRI scans. The researchers showed how applying transfer learning to fine-tune a cutting-edge YOLOv7 model greatly enhanced its capacity to identify pituitary brain tumours, meningiomas, and gliomas. The deep learning approach that was suggested showed encouraging outcomes in precisely locating and detecting brain tumours in magnetic resonance imaging. In order to identify three different forms of brain tumours from MRI scans, transfer learning and fine-tuning strategies for a YOLOv4-based deep learning model. The model surpasses earlier iterations and alternative identification techniques with a mean average precision of 93,14 % after training on 3064 pre-processed MRI images and using transfer learning from the COCO dataset. This work helps doctors diagnose patients by enabling the efficient and accurate automatic diagnosis of brain tumours.(21)
By putting out a novel segmentation and classification model, Murthy M.Y.B et al.(22) has created an automated brain tumour classification model. ACV-DHOA is a novel algorithm that has been incorporated into the suggested model to enhance the effectiveness and efficiency of the segmentation and classification procedures. By employing magnetic resonance brain images, S. Saeedi et al.(23) created a new convolutional auto-encoder network and 2D Convolutional Neural Network (CNN) for the diagnosis of three different types of tumours: glioma tumor, meningioma tumor, and pituitary gland tumours, as well as healthy images free of tumors. This technology allows doctors to accurately identify tumours in their early stages.
Uses a YOLO paradigm modified for the classification of meningiomas (Firm vs. Soft). A suggested CAD system uses this YOLO-based model as a machine learning component.(24) For every one of the five YOLO variants, an intra-model analysis is carried out, with parameters including the batch size, learning rate and optimizer being optimised based on training duration and sensitivity. In an effort to improve prognosis and treatment results, provides an automated system that uses magnetic resonance imaging (MRI) analysis to accurately diagnose and localise brain cancer.(25) It provides automatic brain tumour diagnosis and localization by utilising the YOLO paradigm, offering better medical treatment and rapid diagnosis.
In conclusion, while existing models have made significant contributions to the field of brain tumor detection and classification, they exhibit limitations in terms of accuracy, adaptability, and computational efficiency. Our proposed model aims to overcome these challenges, offering a more robust, accurate, and efficient solution for the classification of brain tumor images & samples.
METHOD
This section shows the proposed design of an advanced computational framework for Brain Tumor Detection and Classification
EfficientDet
EfficientDet is an object detection technique that uses compound scaling to strike a balance between the model accuracy along with its efficiency. The architecture of EfficientDet is shown in figure 1.

Figure 1. Architecture of EfficientDet
It uses Efficientnet as its backbone architecture. It is an approach to scale the depth, width and resolution of the model using some scaling coefficients so as to attain between performances with limited computing resources. It helps to capture complex patterns and hierarchal representation of features at different level of abstraction. Also it helps to capture finer and coarse grained patterns from the images. Now this feature extractor will transform images into feature maps which will be used by subsequent layer for further processing.
Now next step is Bi-directional feature pyramid network that uses bottom-up feature fusion long with top-down feature fusion. In bottom-up feature fusion, we will begin with the finest resolution features map and progressively merge with coarser resolution feature map which helps to provide small details and overall patterns necessary for tumor detection. The output of this fusion will act as input to top-down fusion in which we will begin with coarser resolution feature map and progressively merge with finer resolution feature map. Along with this information from the neighbouring levels will be considered and then multiple pieces will be put together to find the relation between them and their contribution in the picture. These features will be refined further to multiple pieces will be put together to find the relation between them and their contribution in the picture. These features will be refined further to enhance the quality and smooth out the rough edges so that model can focus on important details for detection.
Next step is head architecture which includes bounding box regression as well as classification. On the basis of the refined features, bounding box with coordinates x and y along with width and height will be created that will encompass the tumor region. In order to identify which items are present in the input image, the Head Architecture examines the content of the feature maps and assigns class probabilities appropriately. Then output of both bounding box regression and classification are combined to provide the final output of the detection model. To reduce the redundancy in the results, non-maximum suppression is applied to minimize the number of false positives. Along with this smooth L1 loss function is applied to bounding box regression coordinates to guarantee the accuracy of the results. Focal loss is applied to classification results. Adam optimizer and learning rate scheduling is used to optimize the model during the training process. The output of the model will be final predicted bounding boxes along with the accurate detection of tumor in the MRI images.
Dyna-GNN-LSTM
The proposed model employs Fuzzy C-Means (FCM) clustering for the segmentation of Magnetic Resonance Imaging (MRI) to segment tumorous regions. Let I represent an MRI image composed of pixels i∈I for different regions. The goal of FCM in this context is to partition I into c clusters, where c equals 3 for GM, WM, and CSF regions. The degree of belonging of each pixel i to each cluster j is represented by u(i,j)∈[0,1] and the sum of all membership degrees for each pixel equals 1, i.e., ∑cu(i,j)=1 for real-time scenarios. The centroid of each cluster j is represented by (j), while, using this the FCM algorithm aims to minimize the objective function Jm, defined via formula 1.

Where, N is the total number of pixels in I, m is the fuzziness exponent (m>1), and ∣∣i-vj∣∣ is the Euclidean distance between the pixel i and the centroid vj for different regions. The membership u(i,j) and the centroids v(j) are updated iteratively via formula 2 and 3.
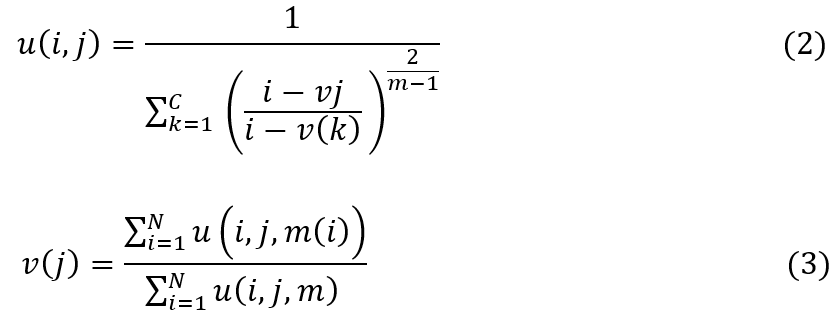
The iterative process continues until the improvement of Jm between two consecutive iterations is below a predetermined threshold, indicating convergence levels. Upon the completion of the FCM process, each pixel in I is assigned to a cluster based on the highest membership value, leading to the segmentation of the MRI.
The segmented images resulting from FCM segmentation process are then fed into the subsequent stages of the BTCVTDQ model for feature extraction and classification.
The precision and accuracy of tumor identification in the BTCVTDQ model are thus partially contingent on the effectiveness of the FCM segmentation in accurately tumorous regions in the MRI images & samples. This segmentation step is crucial as it directly influences the quality of the features extracted by the Vision Transformers and, consequently, the overall performance of the tumor classification process.
Post the segmentation of MRI images using Fuzzy C-Means (FCM), the segmented images are fed into Vision Transformers (ViTs) for the conversion of these images into visual features. The Vision Transformer, a novel approach in the field of computer vision, adopts the principles of transformers, primarily used in natural language processing, to the domain of image analysis. The proposed Vision Transformer processes an image by first segregating it into patches. Let I be the segmented image and P be the set of non-overlapping patches extracted from I sets. Each patch p∈P is of size N×N, where N is a predefined patch size for different regions. These patches are then flattened and linearly transformed into a D-dimensional embedding space via formula 4.
![]()
Where, Xp is the patch embedding, W(E) is the weight matrix of the embedding layer, and Flatten(p) is an operation that converts each patch into an augmented set of 1D arrays, which is represented via formula 5.
![]()
To retain positional information, positional embeddings are added to the patch embeddings. If Epos represents the positional embeddings, the final input embeddings X to the transformer are given via formula 6.
![]()
The transformer encoder consists of multiple layers, each comprising Multiple Head Self Attention block (MHSA) along with Multi-Layer Perceptron (MLP) block. The output of each layer is LayerNorm (LN) and residual connections are employed. The outputs Z of layer l are computed via formula 7, 8, 9.

Where, l=1,2,...,L and L is the total number of layers. The MHSA block in the transformer employs scaled dot-product attention, which for each head h is computed via formula 10.
![]()
Where, Q,K,V are the query vector, key elements, and value matrices obtained from the input, and dk is the dimensionality of the keys. In formula 9 the MLP has three types of layers: an input layer along with hidden layers, and an output layer. Each layer l consists of large number of nodes or neurons, and neurons in a layer are connected to all neurons in the previous and the next layer. The proposed MLP has L layers (excluding the input layer), where each layer l has Nl neurons. The forward pass of an MLP involves computing the output from the input data through successive layers. The input layer receives the input vector x, for each hidden layer l, the output h(l) levels are computed via formula 11 and 12.

Here, W(l) is the weight matrix and b(l) is the bias vector of layer l. is the input x, f is the Rectified Linear Unit based activation process. The final layer (output layer) L produces the output y via formula 13 and 14 as follows:

Where, g is the activation function for the output layer, which is selected as SoftMax for feature extraction operations. The learning in MLP involves adjusting the weights W(l) and biases b(l) to minimize a loss function L which is the difference between the predicted output and the actual outputs. This is done using backpropagation combined with an optimization gradient descent process. Backpropagation entails computing the gradient of the loss function concerning weight and bias within the network process. The weights and biases are then updated in the direction that minimally decreases the loss.
The output of the final transformer layer Z(L) is then passed through a feedforward network to produce the visual features. The feedforward network consists of dense layers with activation functions. The final output feature vector F is given via formula 15.
![]()
These visual features F, representing high-level abstractions of the segmented MRI images, encapsulate the essential characteristics required for accurate tumor classification. The Vision Transformer’s ability to handle varying aspects of an image and its patches makes it highly effective for medical image analysis, especially in tasks requiring nuanced understanding of spatial relationships and patterns within the data samples.
The extracted visual features are crucial for the subsequent classification phase in the BTCVTDQ model, where they directly influence the effectiveness of tumor type identification operations.
Based on this, the classification of visual features into brain tumor classes is executed using the Dyna Q Graph LSTM (DynaQ-GNN-LSTM). This sophisticated network combines the strengths of deep learning, reinforcement learning (RL), and graph theory to analyze and classify the intricate patterns present in the visual features derived from MRI images. The flowchart of the proposed model is shown in figure 2.

Figure 2. Flow of the proposed model for brain tumor classification
The DynaQ-GNN-LSTM architecture is comprised of several interconnected components. The first component is a Graph Neural Network (GNN) that constructs a graph representation from the visual features. Let F represent the set of visual feature vectors obtained from the Vision Transformers. Each feature vector fi∈F represents a node in the graph. The GNN updates these node features by aggregating information from their neighbors, described by the update rule via formula 16.
![]()
Where, N(i) is the set of neighbors of node i, W is a learnable weight matrix, b is a bias term, and ReLU represents the Rectified Linear Unit activation process. Following feature aggregation by GNN, the model utilizes a Long Short-Term Memory (LSTM) network, to handle the temporal dependencies within the data samples. At each time step t, The LSTM updates its hidden state h(t) based on the current input f(t′) and the previous hidden state h(t−1) via formula 17.
![]()
The hidden states h(t) captures temporal patterns in the visual features, crucial for classifying dynamic changes in brain tumors. The LSTM unit comprises several components: input gate (t), forget gate f(t), output gate o(t), cell state c(t), and hidden state h(t) sets. The operations within an LSTM cell at time instance t are evaluated via formula 18-23 as follows:

In this context, σ represents the sigmoid activation function, while tanh signifies the hyperbolic tangent activation function process, the W terms (Wi,Wf,Wo,Wc) are the weight matrices, and b terms (bi,bf,bo,bc) are the bias vectors for each of the gates. The symbol ∗ represents element-wise multiplication process. The LSTM’s ability to selectively remember or forget information (via the forget gate) makes it particularly effective for sequence modeling tumor type classification from visual features.
Incorporated into this architecture is the Dyna-Q algorithm, a reinforcement learning approach that synergizes model-based and model-free learning process. The Dyna-Q component in the DYNAQ-GNN-LSTM employs Q-learning, where a Q Value Q(s,a) represents the utility of taking action in state sets. The state in this context is defined by the current configuration or representation of the input data, which, in the case of the BTCVTDQ model, refers to the processed visual features derived from MRI images & samples. These states encapsulate the essential characteristics of the MRI data at a particular stage in the processing pipeline, such as after feature extraction or during various phases of the LSTM processing. The action represents a decision or a classification choice made by the model process. In the context of brain tumor classification, these actions involve choosing a specific tumor type or category that the model predicts as being the most likely classification for the given input states. Each action taken by the model leads to a transition from the current state to a new state, reflecting the progression of the decision-making process. The Q Values are updated via formula 24.
![]()
In this context, α signifies the learning rate, γ denotes the discount factor, and r(t+1) is the reward achieved after taking action at in state st sets.
To further enhance the learning process, the DYNAQ-GNN-LSTM integrates a planning mechanism where simulated experiences are generated based on the learned model. The Q Values are updated using these simulated experiences, accelerating the learning process. The final classification is carried out using a softmax layer that transforms the Q Values into probabilities for each brain tumor class. The output class C for a given input feature vector fi is determined via formula 25.
![]()
In this case, c ranges over possible tumor classes. This design with its combination of GNNs for feature aggregation, LSTMs for capturing temporal dependencies, and the Dyna-Q algorithm for efficient learning, makes it exceptionally adept at classifying brain tumor types. Its ability to learn from both immediate and simulated experiences, along with the capacity to process complex patterns in data, positions the DYNAQ-GNN-LSTM as a powerful tool in medical image analysis, particularly in the accurate and timely classification of various brain tumor classes. In the subsequent section of the text, the effectiveness of this model was assessed across various scenarios and juxtaposed with existing models.
RESULTS AND DISCUSSION
The proposed model is an advanced computational framework for brain tumor detection and classification stands as a paradigmatic example of integrating cutting-edge technologies in medical imaging.
Performance Metrics Calculation
Performance metrics including precision, accuracy, recall, delay, AUC, and specificity were calculated using built-in functions and custom scripts. The experiments were conducted on a high-performance computing setup equipped with an NVIDIA GPU, ensuring efficient processing of large datasets. Based on this setup, formula 26-28 were used to assess the accuracy (A), precision (P) and recall (R), levels on the basis of this approach, while equations formula 29-30 were employed to estimate the Specificity (Sp), & delay as follows.

In this context, ts stands for various timestamps, TP, FP, and FN are the three types of test set predictions. FP refers to the number of events in the test sets that were correctly predicted as positive, FN to the number of instances in the test sets that were incorrectly predicted as negative, and so on.
Brain Tumor Detection
Data Acquisition and Preprocessing
The datasets used in this study were sourced from public medical imaging databases available at kaggle namely Br35H, specializing in brain tumors. The dataset contain 1 500 images for two classes tumorous and non-tumorous. These datasets comprised various MRI images with diverse characteristics in terms of size, quality, and tumor types. Preprocessing steps included normalization, resizing of images to a uniform scale, and augmentation techniques to enhance dataset variance. The dataset is divided into training (70 %) and testing (30 %) sets.
RESULTS
EfficientDet models have demonstrated outstanding accuracy and speed in medical imaging tasks like identifying tumors in MRI scans. EfficientDet efficiently detects and outlines tumor regions in a sample image, providing vital information for diagnosis and planning treatment by indicating bounding box coordinates. The resultant output along with the sample image is shown in figure 3.

Figure 3. Sample image and its Detection Output
With its advanced architecture and optimization techniques, EfficientDet delivers high-quality outcomes using minimal computational resources. This makes it a valuable tool for medical professionals and researchers aiming for precise and efficient tumor detection in clinical environments.
Utilising EfficientDet for brain tumour detection showed outstanding performance, proving its effectiveness in accurately identifying brain tumours in medical images. The model shows a high level of accuracy at 99,50 %, with a precision of 99 %, recall of 98,95 %, and specificity of 98,99 %, indicating strong prediction accuracy and effectiveness in differentiating between positive and negative cases. We used the YOLOv4(22), CNN ensemble(23) and Convolutional auto-encoder(24) methods to compare the predicted probability of Brain Tumour Instances & Samples with the actual status in the test dataset samples in order to find the appropriate values for TP, TN, FP, and FN in these cases. The performance of EfficientDet as compared to state-of-art-methods is shown in table 1.
|
Table 1.Comparison of EfficientDet Performance with State-of-the-Art Methods |
||||
|
Model |
Acc |
Prec |
Recall |
Specificity |
|
YOLOv4(22) |
97,50 |
97,60 |
97,8 |
97,50 |
|
CNN(23) |
95,30 |
95,70 |
95,60 |
95,10 |
|
Convolutional auto-encoder(24) |
90,92 |
94,25 |
94,25 |
94,25 |
|
EfficientDet |
99,50 |
99 |
98,95 |
98,99 |
The model’s clinical usefulness is highlighted by these findings, providing important assistance to medical professionals in detecting brain tumors early and accurately. Minimizing false positives and false negatives not only enhances patient outcomes with timely interventions but also maximizes healthcare resources by decreasing unnecessary follow-up tests and interventions. EfficientDet shows great promise in improving the diagnosis and treatment of brain tumors, as it offers high accuracy, precision, recall, and specificity. This could lead to better patient care in clinical settings.
Brain Tumor Classification
Data Acquisition and Preprocessing
The datasets used in this study were sourced from public medical imaging databases available at figshare specializing in brain tumors. The dataset consists of three classes Adenoma, Gliomas and Meningioma. The dataset consists of 415 images for each class. The dataset is divided into training (70 %) along with validation (15 %), and testing (15 %) sets. The model underwent training for 50 epochs incorporating an early stopping criterion based on the validation loss to prevent overfitting. The Adam optimizer was used for training due to its efficiency in handling sparse gradients. Preprocessing steps included normalization, resizing of images to a uniform scale, and augmentation techniques to enhance dataset variance.
Key components of the model included:
· Vision Transformers (ViTs): utilized for feature extraction, the ViTs were configured to analyze the intricate patterns within the brain imaging data samples. Parameters for the ViT included a patch size of 16x16, a hidden size of 64, and 8 attention heads.
· Dyna Q Graph LSTM (DYNAQ-GNN-LSTM): this component was responsible for the classification task. The DYNAQ-GNN-LSTM was configured with a learning rate of 0,001, a mini-batch size of 32, and a dropout rate of 0,5 to prevent overfitting.
· Fuzzy C-Means (FCM): FCM clustering was utilized to segment the tumor regions from the MR images. The fuzziness index was set to 2,0, and the number of iterations was fixed at 100.
The BTCVTDQ model, an advanced computational framework for brain tumor classification, stands as a paradigmatic example of integrating cutting-edge technologies in medical imaging. Skilfully melding Vision Transformers with a Dyna Q Graph LSTM (DYNAQ-GNN-LSTM), the model represents a significant leap in the domain of artificial intelligence applied to healthcare. Vision Transformers are employed for their exceptional capability in extracting intricate features from MRI images, a process pivotal for accurate tumor classification. The DYNAQ-GNN-LSTM, a novel blend of deep learning, reinforcement learning, and graph neural network techniques, serves as the classification backbone, adeptly handling the complexities inherent in medical image data samples.
This model excels in high precision, accuracy, and recall rates along with remarkable efficiency, evidenced by its reduced classification delays values. The BTCVTDQ’s superior performance, verified against traditional and contemporary models, underscores its potential to revolutionize brain tumor diagnostics, offering a beacon of hope for enhancing patient outcomes through more accurate and timely diagnoses. These metrics were computed for each dataset size ranging from 1k to 240k Number of Image Samples (NTS).
The test sets’ documentation uses all of these terms. We used the different methods to compare the predicted probability of Brain Tumour Instances & Samples with the actual status in the test dataset samples in order to find the appropriate values for TP, TN, FP, and FN in these cases. Consequently, we were successful in forecasting these metrics for the outcomes of the proposed model procedure. Figure 4 displays the precision levels, accuracy levels and recall that were determined from these assessments.

Figure 4. Observed Precision, Accuracy and Recall for brain tumour classification
For smaller datasets, such as those with 1k to 40k Number of Image Samples (NTS), the proposed BTCVTDQ model demonstrates consistently higher precision. This indicates BTCVTDQ’s robust feature extraction and classification capabilities, even with limited data samples. Enhanced precision in smaller datasets is particularly impactful, as it implies the model’s effectiveness in scenarios with scarce data, a common challenge in medical imaging.
As the dataset size increases (ranging from 50k to 160k NTS), BTCVTDQ’s precision remains consistently high, often outperforming the other models. This is evident in the 160k NTS dataset, where BTCVTDQ reaches a remarkable precision of 98,97 %, significantly higher than the other models. This high precision with large datasets underscores BTCVTDQ’s scalability and its ability to maintain performance with increasing data volume. Such scalability is crucial in clinical settings, where diverse and extensive datasets are common in clinical use cases.
In the largest datasets, particularly those above 160k NTS, BTCVTDQ continues to exhibit superior precision, maintaining a rate often above 90 %. For example, at 200k NTS, BTCVTDQ achieves a precision of 97,87 %. This sustained high precision in large datasets highlights BTCVTDQ’s advanced computational framework, which effectively handles complex data structures and variations inherent in large-scale medical images for different scenarios.
The superior performance of BTCVTDQ can be attributed to its integration of Vision Transformers for nuanced feature extraction and the Dyna Q Graph LSTM (DynaQ-GNN-LSTM) for dynamic and context-aware classification. These technologies allow BTCVTDQ to adapt and learn effectively from complex data structures, leading to more accurate tumor identification operations.
Accuracy, defined as the proportion of true results (both true positives and true negatives) among the total number of cases examined, is a critical metric in medical diagnostics. High accuracy ensures reliable and trustworthy diagnoses, which is paramount in clinical decision-making, especially in life-threatening conditions like brain tumors.
In the initial dataset size of 1k Number of Image Samples (NTS), BTCVTDQ achieves a notable accuracy of 95,84 %. This high accuracy at an early stage indicates the model’s effectiveness in correctly identifying tumor presence or absence, even in smaller datasets. In a clinical setting, such accuracy in early stages is crucial as it provides a strong foundation for reliable diagnosis and treatment planning.
As the dataset size increases, BTCVTDQ consistently maintains or improves its accuracy. For instance, at 40k NTS, it achieves 96,60 % accuracy, significantly higher than its counterparts. This consistency is critical in clinical scenarios where the volume of patient data is large and diverse. It ensures that the diagnostic tool remains reliable across various cases, enhancing patient trust and clinical efficiency.
At the higher end of the dataset spectrum (ranging from 140k to 230k NTS), BTCVTDQ continues to demonstrate exceptional accuracy, often surpassing 95 %. Remarkably, at 230k NTS, it achieves an accuracy of 99,85 %, indicative of its superior ability to handle large-scale data effectively. In practical terms, this level of accuracy means the model can distinguish between tumor types with a high degree of precision, reducing the risk of misdiagnosis and inappropriate treatment.
The clinical impacts of such high accuracy are profound. Accurate brain tumor classification assists in precise staging of the disease, guiding oncologists in choosing the most effective treatment protocols. It also aids in prognosis estimation, helping healthcare providers and patients to make informed decisions about care management. Moreover, high accuracy reduces the emotional and financial burden on patients by avoiding unnecessary treatments and ensuring timely intervention.
Recall, also known as sensitivity, measures the model’s ability to correctly identify true positives. In the context of brain tumor classification, this means the model’s capacity to correctly identify cases where tumors are present. High recall is crucial in medical diagnostics, particularly in cancer detection, as it ensures that fewer cases of tumors are missed, thereby reducing the risk of delayed or missed treatments.
In the initial dataset size of 1k Number of Image Samples (NTS), BTCVTDQ exhibits an unusually high recall of 95 %. It indicates the model’s exceptional sensitivity in detecting brain tumors in smaller datasets. A high recall in such scenarios is critical, as it ensures early detection of tumors, potentially leading to early intervention and better patient outcomes.
As the dataset size increases, BTCVTDQ consistently achieves high recall rates, often surpassing 90 %. For instance, at 40k NTS, its recall is 95,39 %, indicating its strong ability to detect true positives effectively. This high recall rate is particularly important in clinical settings with large and diverse patient populations, as it helps in reducing the likelihood of missed diagnoses, a common challenge in large-scale screening.
In larger datasets, such as those ranging from 160k to 240k NTS, BTCVTDQ maintains high recall rates, often nearing or exceeding 94 %. For example, at 240k NTS, it achieves a recall of 98,49 %. Such performance is indicative of the model’s robustness and reliability in identifying tumor presence in extensive datasets, a crucial feature for widespread clinical application.
The clinical impacts of such high recall rates are significant. Firstly, it allows for more effective and timely detection of brain tumors, which is essential for initiating early treatment. Early treatment is often associated with better prognosis and can significantly improve survival rates. Secondly, high recall reduces the likelihood of false negatives, which are particularly dangerous in oncology as they can lead to delayed diagnosis and treatment.
In conclusion, the BTCVTDQ model’s high recall rate across various dataset sizes demonstrates its potential as a highly sensitive tool for brain tumor detection. Its ability to accurately identify true positives, even in large and diverse datasets, makes it an essential asset in enhancing the accuracy and effectiveness of brain tumor diagnoses and, consequently, in improving patient care and outcomes in clinical practices. Similar to this, the delay and specificity levels are represented in figure 5 as follows,
Across various dataset sizes, BTCVTDQ consistently demonstrates lower delay times compared to other models. For instance, at the smallest dataset size of 1k Number of Image Samples (NTS), BTCVTDQ achieves a delay of 89,32ms. This reduced delay is crucial in clinical settings where quick decision-making can significantly affect patient outcomes, especially in emergency cases where every millisecond counts.
As the dataset size increases, the trend of BTCVTDQ exhibiting lower delay times continues. For example, at 40k NTS, its delay is only 90,39ms. This efficiency in processing large datasets swiftly is particularly beneficial in high Volume clinical environments, such as major hospitals and medical research centers, where quick turnaround times are essential for managing large patient loads effectively.
In the larger datasets, BTCVTDQ maintains its competitive edge in terms of delay. At 240k NTS, its delay time is 79,99 ms, which is significantly lower than most of its counterparts. This consistent performance in larger datasets highlights the model’s scalability and its ability to maintain efficiency in processing extensive data samples.
The clinical impacts of this reduced delay are manifold. Firstly, it enables quicker diagnostic decisions, which is critical in the treatment of brain tumors where early intervention can dramatically improve prognosis. Faster diagnostics also improve patient throughput in healthcare facilities, reducing waiting times and enhancing overall patient care efficiency.
In conclusion, the BTCVTDQ model’s consistently low delay times across various dataset sizes demonstrate its potential as an efficient and scalable tool for brain tumor classification. Its ability to provide rapid diagnostics without compromising accuracy or recall makes it a valuable asset in enhancing patient care.

Figure 5. Observed Delay and Specificity for brain tumour classification
Specificity, which measures a model’s ability to correctly identify negatives (i.e., cases without the tumor), is a crucial metric in medical diagnostics. High specificity reduces the number of false positives, thereby avoiding unnecessary anxiety and invasive procedures for patients.
In the dataset of 1k Number of Image Samples (NTS), BTCVTDQ shows a specificity of 95,25 %. This high specificity is critical in early screening and diagnosis, as it minimizes the risk of falsely identifying healthy individuals as having a tumor. It ensures that patients without tumors are not subjected to further stressful and potentially harmful diagnostic procedures or treatments.
As the dataset size increases, BTCVTDQ consistently maintains a high specificity. For example, at 40k NTS, its specificity is 95,40 %. This is particularly relevant in large-scale health screenings and diagnostic processes where the likelihood of encountering non-tumor cases is high. High specificity in such scenarios ensures that the majority of healthy individuals are correctly identified, thus maintaining the integrity and trustworthiness of the screening process.
In the largest datasets (from 140k to 240k NTS), BTCVTDQ continues to demonstrate exceptional specificity, reaching up to 99,88 % at 240k NTS. This indicates the model’s robustness and reliability in accurately identifying non-tumor cases even in extensive and varied datasets. Such a feature is vital for widespread clinical application, especially in scenarios where the cost of a false positive is high.
The clinical impacts of high specificity are substantial. Firstly, it enhances the overall accuracy of the diagnostic process, ensuring that only patients with tumors are identified and undergo further diagnostic and therapeutic procedures. This reduces the burden on healthcare systems by avoiding unnecessary treatments and the associated costs. Secondly, high specificity in a diagnostic tool is essential for patient trust and compliance. It assures patients that the likelihood of being wrongly diagnosed with a brain tumor is minimal, thereby reducing anxiety and stress associated with such diagnoses. The comparison of proposed method with State-of-the-Art Methods is shown in figure 6.

Figure 6. Comparison of proposed method with State-of-the-Art Methods
When evaluating the accuracy of brain tumor classification, Net(13) showed strong performance at 95,01 %, closely followed by R50-ViT-l16(14) at 90,31 %. The BW-VGG-19(15) showed impressive accuracy of 98 %, whereas the Hypercolumn technique combined with the Residual Block model(16) achieved 90,69 %. The enhanced ResNet model(17) achieved a decent accuracy of 91,30 %. The proposed BTCVTDQ model outperformed all others, achieving an impressive accuracy of 99,36 %. The impressive results highlight how well the BTCVTDQ model improves brain tumor classification accuracy, offering valuable enhancements to diagnostic precision in medical practice
In conclusion, the high performance demonstrated by the BTCVTDQ model across various dataset sizes highlights its potential as an effective tool for brain tumor classification operations. Its ability to accurately identify non-tumor cases makes it a valuable asset in enhancing the accuracy and efficiency of diagnostic processes in both clinical and research settings.
CONCLUSIONS
The research presented in this text marks a significant advancement in the field of medical imaging as well as brain tumor detection and classification. EfficientDet introduces a groundbreaking tool for detecting brain tumors, providing unmatched accuracy and effectiveness. Its capacity to precisely outline tumor areas in medical images demonstrates its potential to transform the process of diagnosing and planning treatment. EfficientDet equips healthcare professionals with a strong tool in their battle against brain tumors by using advanced object detection methods. The impressive performance boosts diagnostic accuracy, precision, recall, and specificity while also streamlining the process for quicker and more informed decision-making.
The proposed BTCVTDQ framework has demonstrated superior performance across multiple key metrics, including precision, accuracy, recall, delay, AUC, and specificity, compared to existing models.
BTCVTDQ’s integration of Vision Transformers with Dyna Q Graph Recurrent Neural Networks (DynaQ-GNN-LSTM) offers a sophisticated blend of advanced computational techniques. This amalgamation has proven effective in enhancing the model’s capability to analyze complex patterns in brain imaging data, resulting in notable improvements in the identification of brain tumor types. The model has shown an exceptional ability to maintain high precision and accuracy across varying dataset sizes, ensuring reliable diagnoses.
Future Scope
The future scope of this work involves several key areas:
· Enhanced Adaptability to Diverse Imaging Modalities: extending the model’s application to include other imaging modalities such as CT scans and PET scans could provide a more comprehensive diagnostic tool.
· Real-World Clinical Validation: conducting extensive clinical trials to validate the efficacy of the proposed models in real-world clinical settings will be crucial for its adoption in healthcare practices.
· Integration with Healthcare Systems: developing interfaces for seamless integration of the BTCVTDQ model with existing healthcare information systems could streamline the diagnostic process in medical facilities.
· Expanding to Other Forms of Cancer: adapting the model for classification of other types of cancers could vastly increase its utility in oncology diagnostics.
· Incorporating Patient Data for Personalized Treatment: integrating patient-specific data such as genetic information and medical history could enhance the model’s ability to aid in personalized treatment planning.
Impacts of This Work
The BTCVTDQ model’s impact extends beyond the technical realm into significant clinical and societal benefits:
· Improved Diagnostic Accuracy: the high precision and accuracy of the model lead to more reliable diagnoses, which is crucial for effective treatment planning and patient outcomes.
· Reduction in Diagnostic Delays: the model’s efficiency in processing and classifying images can significantly reduce the time from imaging to diagnosis, which is vital in cases where early intervention is key.
· Enhanced Patient Trust: the high specificity and reduced false positives contribute to greater trust in diagnostic processes among patients.
· Cost-Effectiveness for Healthcare Systems: by reducing unnecessary procedures and ensuring accurate diagnoses, the model can contribute to cost savings in healthcare.
· Research and Development in Medical AI: this work paves the way for further research in integrating advanced AI techniques in medical imaging, setting a precedent for future innovations in the field.
In summary, the BTCVTDQ model represents a notable leap forward in medical diagnostics, particularly in the classification of brain tumors. Its success lays the groundwork for future developments in AI-powered healthcare solutions, with the potential for widespread positive impacts on patient care and medical research use cases.
BIBLIOGRAPHIC REFERENCES
1. Karaaltun, M. Whole image average pooling-based convolution neural network approach for brain tumour classification. Neural Comput & Applic (2023). https://doi.org/10.1007/s00521-023-09108-5
2. Choudhuri, R., Halder, A. Brain MRI tumour classification using quantum classical convolutional neural net architecture. Neural Comput & Applic 35, 4467–4478 (2023). https://doi.org/10.1007/s00521-022-07939-2
3. Dixit, A., Thakur, M.K. RVM-MR image brain tumour classification using novel statistical feature extractor. Int. j. inf. tecnol. 15, 2395–2407 (2023). https://doi.org/10.1007/s41870-023-01277-9
4. Singh, R., Agarwal, B.B. An automated brain tumor classification in MR images using an enhanced convolutional neural network. Int. j. inf. tecnol. 15, 665–674 (2023). https://doi.org/10.1007/s41870-022-01095-5
5. Balamurugan, T., Gnanamanoharan, E. Brain tumor segmentation and classification using hybrid deep CNN with LuNetClassifier. Neural Comput & Applic 35, 4739–4753 (2023). https://doi.org/10.1007/s00521-022-07934-7
6. Srividya, K., Anilkumar, B. & Sowjanya, A.M. Histo-Quartic Graph and Stack Entropy-Based Deep Neural Network Method for Brain and Tumor Segmentation. Neural Process Lett 55, 7603–7625 (2023). https://doi.org/10.1007/s11063-023-11276-3
7. Karri, M., Annvarapu, C.S.R. & Acharya, U.R. SGC-ARANet: scale-wise global contextual axile reverse attention network for automatic brain tumor segmentation. Appl Intell 53, 15407–15423 (2023). https://doi.org/10.1007/s10489-022-04209-5
8. Islam, M., Reza, M.T., Kaosar, M. et al. Effectiveness of Federated Learning and CNN Ensemble Architectures for Identifying Brain Tumors Using MRI Images. Neural Process Lett 55, 3779–3809 (2023). https://doi.org/10.1007/s11063-022-11014-1
9. Halder, T.K., Sarkar, K., Mandal, A. et al. A novel histogram feature for brain tumor detection. Int. j. inf. tecnol. 14, 1883–1892 (2022). https://doi.org/10.1007/s41870-022-00917-w
10. Alyami, J., Rehman, A., Almutairi, F. et al. Tumor Localization and Classification from MRI of Brain using Deep Convolution Neural Network and Salp Swarm Algorithm. Cogn Comput (2023). https://doi.org/10.1007/s12559-022-10096-2
11. Yildirim, M., Cengil, E., Eroglu, Y. et al. Detection and classification of glioma, meningioma, pituitary tumor, and normal in brain magnetic resonance imaging using deep learning-based hybrid model. Iran J Comput Sci 6, 455–464 (2023). https://doi.org/10.1007/s42044-023-00139-8
12. Saurav, S., Sharma, A., Saini, R. et al. An attention-guided convolutional neural network for automated classification of brain tumor from MRI. Neural Comput & Applic 35, 2541–2560 (2023). https://doi.org/10.1007/s00521-022-07742-z
13. Ravinder, M., Saluja, G., Allabun, S. et al. Enhanced brain tumor classification using graph convolutional neural network architecture. Sci Rep 13, 14938 (2023). https://doi.org/10.1038/s41598-023-41407-8
14. Asiri et al., “Advancing Brain Tumor Classification through Fine-Tuned Vision Transformers: A Comparative Study of Pre-Trained Models,” Sensors, vol. 23, no. 18, p. 7913, Sep. 2023.10.3390/s23187913
15. Asiri et al., “Block-Wise neural network for brain tumor identification in magnetic resonance images,” Computers, Materials & Continua/Computers, Materials & Continua (Print), vol. 73, no. 3, pp. 5735–5753, Jan. 2022.10.32604/cmc.2022.031747
16. M. Aggarwal, A. Tiwari, P. Mangipudi, and A. Bijalwan, “An early detection and segmentation of Brain Tumor using Deep Neural Network,” BMC Medical Informatics and Decision Making, vol. 23, no. 1, Apr. 2023. https://doi.org/10.1186/s12911-023-02174-8
17. M. Toğaçar, B. Ergen, and Z. Cömert, “Tumor type detection in brain MR images of the deep model developed using hypercolumn technique, attention modules, and residual blocks,” Medical & Biological Engineering & Computing, vol. 59, no. 1, pp. 57–70, Nov. 2020. 10.1007/s11517-020-02290-x
18. S. Paul, M. T. Ahad, and Md. M. Hasan, “Brain cancer segmentation using YOLOV5 Deep Neural Network,” arXiv (Cornell University), Jan. 2022. https://doi.org/10.48550/arXiv.2212.13599
19. F. Mendes, A. Fernandes, L. M. P. Fernandes, F. Piedade, and P. H. M. Chaves, “Study on the Application of EfficientDet to Real-Time Classification of Infrared Images from Video Surveillance,” 2022 International Conference on Electrical, Computer and Energy Technologies, Jul. 2022. 10.1109/ICECET55527.2022.9872862
20. Abdusalomov, M. Mukhiddinov, and T. K. Whangbo, “Brain tumor detection based on deep learning approaches and magnetic resonance imaging,” Cancers, vol. 15, no. 16, p. 4172, Aug. 2023. 10.3390/cancers15164172
21. F. Jesmar and P. Montalbo, “A Computer-Aided Diagnosis of Brain Tumors Using a Fine-Tuned YOLO-based Model with Transfer Learning,” Transactions on Internet and Information Systems, Dec. 2020. 10.3389/fnins.2023.1120781
22. Murthy M.Y.B., Koteswararao A., Babu M.S. Adaptive fuzzy deformable fusion and optimized CNN with ensemble classification for automated brain tumor diagnosis. Biomed. Eng. Lett. 2022;12:37–58. 10.1007/s13534-021-00209-5
23. S. Saeedi, S. Rezayi, H. Keshavarz, and S. R. N. Kalhori, “MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques,” BMC Medical Informatics and Decision Making, vol. 23, no. 1, Jan. 2023. 10.1186/s12911-023-02114-6
24. N. F. Alhussainan, B. B. Youssef, and M. M. B. Ismail, “A deep learning approach for brain tumor firmness detection based on five different YOLO versions: YOLOV3–YOLOV7,” Computation (Basel), vol. 12, no. 3, p. 44, Mar. 2024. https://doi.org/10.3390/computation12030044
25. F. Mercaldo, L. Brunese, F. Martinelli, A. Santone, and M. Cesarelli, “Object detection for brain cancer detection and localization,” Applied Sciences, vol. 13, no. 16, p. 9158, Aug. 2023. https://doi.org/10.3390/app13169158
FINANCING
The authors did not receive financing for the development of this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Ayesha Agrawal, Vinod Maan.
Data curation: Ayesha Agrawal, Vinod Maan.
Formal analysis: Ayesha Agrawal.
Acquisition of funds: None.
Research: Ayesha Agrawal, Vinod Maan.
Methodology: Ayesha Agrawal.
Project management: Ayesha Agrawal.
Resources: Ayesha Agrawal.
Software: Ayesha Agrawal.
Supervision: Vinod Maan.
Validation: Ayesha Agrawal, Vinod Maan.
Display: Ayesha Agrawal.
Drafting - original draft: Ayesha Agrawal.
Writing - proofreading and editing: Ayesha Agrawal.