REVIEW
Emerging Technologies in Industrial Microbiology: From Bioengineering to CRISPR-Cas Systems
Tecnologías emergentes en microbiología industrial: De la bioingeniería a los sistemas CRISPR-Cas
Umar Farooq1 *, Malathi
Hanumanthayya2 ![]() ,
Izharul Haq3
,
Izharul Haq3 ![]()
1Teerthanker Mahaveer University. Department of Microbiology. Moradabad, Uttar Pradesh, India.
2Department of Life Science. School of Sciences, JAIN (Deemed-to-be University). Bangalore, India.
3Assistant Professor. School of Life and Basic Sciences, Jaipur National University. Jaipur, India.
Cite as: Farooq U, Hanumanthayya M, Haq I. Tecnologías emergentes en microbiología industrial: De la bioingeniería a los sistemas CRISPR-Cas. Salud, Ciencia y Tecnología. 2023; 3:406. https://doi.org/10.56294/saludcyt2023406
Received: 09-04-2022 Reviewed: 30-04-2022 Accepted: 12-06-2022 Published: 13-06-2022
Editor: Fasi Ahamad Shaik
ABSTRACT
Synthetic biology advancements have led to the emergence of “engineering biology” via equivalence and platform base on hierarchical, orthogonal, and modularized biological system. Utilizing bioparts found in sequence databases, genome engineering (GE) is required to create synthetic cells with the appropriate functionality. The CRISPR system, one of several tools, is modularly made up of Cas nuclease and guide RNA, making it simple to modify the GE at will. To correctly modify the GE at the distinct nucleotide level, many techniques have recently been devised. The CRISPR-Cas (CC) system has also been expanded to molecular diagnostics for nucleic acids and pathogen identification, together with viruses that cause illness. Furthermore, metabolic biotechnology is expanding to find the target of CC methodology, which can exactly regulate the production of certain genes in cells. In the present article, we evaluate the current state of several CC technology that may be used in synthetic biology and talk about how artificial biology and CC technology have evolved in the field of microbiology.
Keywords: CRISPR-Cas Methodologies; Synthetic Biology; Biotechnology; Nucleic Acid Diagnostics; TransCleavage.
RESUMEN
Los avances de la biología sintética han propiciado la aparición de la “biología de ingeniería” mediante la equivalencia y la plataforma basada en un sistema biológico jerárquico, ortogonal y modularizado. Utilizando biopartes encontradas en bases de datos de secuencias, la ingeniería genómica (GE) es necesaria para crear células sintéticas con la funcionalidad adecuada. El sistema CRISPR, una de las diversas herramientas, está formado modularmente por la nucleasa Cas y el ARN guía, lo que simplifica la modificación de la GE a voluntad. Para modificar correctamente la GE a nivel de nucleótidos distintos, se han ideado recientemente muchas técnicas. El sistema CRISPR-Cas (CC) también se ha extendido al diagnóstico molecular de ácidos nucleicos y a la identificación de patógenos y virus causantes de enfermedades. Además, la biotecnología metabólica se está expandiendo para encontrar el objetivo de la metodología CC, que puede regular exactamente la producción de determinados genes en las células. En el presente artículo, evaluamos el estado actual de varias tecnologías de CC que pueden utilizarse en biología sintética y hablamos de cómo han evolucionado la biología artificial y la tecnología de CC en el campo de la microbiología.
Palabras Clave: Metodologías CRISPR-Cas; Biología Sintética; Biotecnología; Diagnóstico de Ácidos Nucleicos; Trans Cleavage.
INTRODUCTION
Diverse genetic and metabolic capabilities: Non-conventional yeasts have diverse genetic and metabolic capabilities that enable them to perform unique functions such as the degradation of complex substrates, the production of high-value compounds, and the adaptation to extreme environments.(1)
The analogy of evolution as a tinkerer rather than an engineer highlights the fact that evolution does not work towards an end goal or a specific design, but rather produces a wide range of solutions to problems that are often cobbled together from existing parts. This is because evolution operates on existing variations within a population, rather than creating new variations from scratch.(2)
Mycotoxin contamination can cause similar health problems, as well as reduced growth rates, decreased reproductive performance, and increased susceptibility to infectious diseases. Livestock and poultry that consume mycotoxin-contaminated feed can also pass on the mycotoxins to humans through the consumption of contaminated meat, milk, and eggs.(3)
The basic biology of CC systems, including their mechanisms of action, their regulation, and their interactions with other cellular processes. This will not only deepen our understanding of these fascinating systems, but it will also enable the improvement of extra precise and effective GE suppression utensils.(4)
It is crucial to have both a thorough strategy that covers all possible causes of contamination and a strong, focused biomarker for monitoring sanitary conditions in order to avoid contamination.(5) These bacteria are commonly establish in the gastrointestinal tract of human and natural world, and their attendance in food or on food contact surfaces is an indication of poor hygiene and potential fecal contamination. The presence of these bacteria in food can be an indication of inadequate processing, poor handling, or unsanitary conditions.(6)
In microbes, the CC organization has an extensive collection of potential applications, including the development of new antibiotics, the construction of biofuels and other valuable chemicals, and the study of gene function and regulation. CC-based approaches can also be used to engineer microbes with desirable traits, such as increased resistance to environmental stresses or improved production of specific metabolites.(7)
The conventional techniques for changing fungi's genetic makeup take a long time and are laborious. The occupied GE sequences of numerous technologically important filamentous fungus are now publicly available, and CC technology has gained attention for its efficacy in creating modified strains of filamentous fungi.(8)
Endogenous CC-based prokaryotic engineering has a significant deal of promise due to the widespread natural presence of CC systems in most archaea and most bacteria. Once the endogenous CC systems are recognized and defined as functioning in their native hosts, type I systems, which make up the most numerous and varied category, would be repurposed as genetic modification tools.(9) In order to serve as a guide for choosing diverse CC apparatus, we compare the properties of several CC/Cas systems and briefly characterize their processes.(10)
CC/Cas systems are a type of genetic engineering tool that allow scientists to make precise changes to the DNA of living organisms. CC/Cas systems use RNA molecules and enzymes to target specific sequences of DNA and cut them with high accuracy. This technology has revolutionized genetic engineering by making it easier, faster, and cheaper to edit the DNA of organisms, including humans.(11)
The proliferating use of CC-based technology for prokaryotics manufacturing has shown accurate, effective, and scalable GE restriction and tunable transcriptional regulation, which shall be translated into the creation of Probiotic lactobacilli with improved robustness and designer functionalities in the next generation.(12)
A reliable, repeatable process that modifies the GE with accuracy and predictability. System biotechnology and synthetic biology techniques, together with CC technology, may transform how cell factories function in the future by enabling them to have certain novel traits that are not present in nature.(13)
Advancements in genetic engineering techniques have enabled scientists to modify the metabolic pathways of these microorganisms to improve their productivity and yield of target chemicals. This has led to the development of more efficient and cost-effective methods for the production of biofuels and bulk chemicals, which have the potential to be more sustainable and environmentally friendly than traditional methods of production.(14)
The article reviewed that the plan that was gained for using the CC/Cas system to modify and engineer bacteria's and yeast's metabolism. The article focused on the utilization produce tiny molecules of microbial system that is crucial in both chemical and biological processes.(15) The Research summarized the mechanism control of conventional methods of genetic modification by using CC technological advances.(16)
The Essay determined the increasing accurate, effective, adaptable as well as both of which can be used to create probiotic lactobacilli of future generations with improved robustness and designer functionalities.(17) The case study reviewed that the significant developments in CCsystem adaptation, modification and intervention/activation.(18)
The Research reviewed to highlight the opportunities and difficulties of creating dynamically controlled by combining study of the most recent research with examples from the own work.(19) The overview objective of the article demonstrated that environment may allow for more effective GE engineering, which raises the possibility that may interfere with GE editing. The findings may provide insight into how to use and enhance CC systems in different microbes for practices of metabolic engineering and GE editing.(20)
The Research summarized the current state of different CC techniques that can be used in imitation ecology, as well as how synthetic biology and CC technology have evolved in the field of microbiology.(21) The article has discussed the promise and numerous applications in the synthesis of heterogonous proteins. In the essay, the mechanics underlying this metabolic engineering strategy are discussed, and its various applications in biotechnology and protein production are highlighted.(22)
The article acted as a guide for the use of this technology in the study of the functions of the GE, in synthetic biology, in genetic advancement, and in the development of new medicinal plant germplasm. In the near future, CC is anticipated revolutionising the use of therapeutic plants in biotechnology.(23) The essay discussed the major turning points in the history of CC/Cas GE suppression equipment, from its original discovery to the present-day developments, including potential medicinal uses.(24)
The research determines how well-known novel technologies including extreme pressure manufacturing, vibrated electricity, ultrasounds, oxygen manufacturing, ultraviolet radiation, a magnetic field, radiation from microwaves, non-thermal the bloodstream electrolyzed deteriorating water, and plasma therapy activated water affect the development and physical traits of various species of seeds. The causes of these technologies' beneficial and detrimental effects on germination rate have previously been examined.(25)
In this study we will discuss about Cas nuclease and guide RNA in the CRISPR (CC) technology make GE modification easy. Recently developed methods change the GE at the nucleotide level. The CC-Cas (CC) technology now detects nucleic acids, pathogens, and viruses that cause sickness.
DEVELOPMENT
CC GE Editing
RNA, which has the two components of the CC system, which exhibit adaptive resistance in bacteria. A lot of species, including bacteria and yeasts, employ it as a tool for GE editing since the objective nucleotide progression may be flexibly changed based on the change in the RNA sequence. Table 1 provide an indication of research on CCGE editing studies in a variety of microorganisms, including bacteria, yeasts, and archaea.
Escherichia coli were the first bacteria for which CC-Cas9-mediate gene edits was made public. We perform gene deleted, insertions, and base substitutions in Staphylococcus aureus using specific plasma including Cas9, sgRNA, -Red recombinased, & donor DNA. Very elevated homologous recombinased efficiency (> 93 percent) is reached when the donor DNA’ arm is 210 base pairs in length in a flamentous fungus. Up to tri heterologous genes may be concurrently introduced into assorted region of the E utilising the C-Cpf1 and Red recombinase.
Using metagenomic data, Cas12f1, which has a gene that is smaller than those of Cas9 and Cpf1, was recently discovered. Additionally, investigation into the origins of the Cas9 and Cpf1 nucleases resulted in the identification of a novel family of RNA-guides nucleases that includes the IscB and TnpB enzymes. Isc B and Tnp B, which are thought to be Cas9 and Cas12 nucleases ancestors, contain a inferior editing efficiency. The protein's small dimension is an advantage when utilised for gene therapy.
|
Table 1. Microbiological pathogens: CC-based diagnostics |
|||||
|
Target amplifcation |
Chromophores/ fuorophores |
Target pathogen |
Limit of Detection |
Cas effector |
Sample type |
|
Reverse transcription, EXPAR |
Fluorescence (SYBR Green I) |
Listeria monocytogenes |
0,82 amole |
Cas9 |
Total RNA |
|
PCR |
Luminescence (Firefy luciferase) |
M. tuberculosi |
50 pM* |
dCas9 |
gDNA |
|
PCR |
Fluorescence (HEX) |
“Pseudorabies virus” (PRV), “Japanese encephalitis virus” (JEV) |
0,5 aM |
Cas12a |
gDNA |
|
LAMP, Asymmetric PCR |
Fluorescence (FAM) |
JEV |
10 aM F |
Cas12b |
Virus |
Identifying Target Sequence
It was discovered that removing similar sequences beyond the target had an target impact when CC-mediated GE editing was studied in the eukaryotic system, which has a very complex GE. It is known that under some conditions, cleavage occurs in unfavorable locations in the GE, posing a challenge to editing. Studies have been done to solve this problem by creating guide RNA or Cas nuclease in eukaryotic cells to enlarge intention specificity and editing effectiveness.
The consistency and modifying effectiveness of the crRNA are improved by Methylation and fluorination, two chemical changes of the Cpf1 crRNA terminal. Once a piece of the separator in RNA is replace with “DNA” the edited competence is improves and the emphasize impact is compact via changes the object's required force.
Cas protein editing is a new approach to improve target specificity. For instance, pairing a Cas9 with a chromatin-modulating peptide may improve on-target activity. In a separate study, nicks are made at various locations and strands using deactivated Cas9 and the FokI nuclease, which reduces the target impact. With a 100 % chance of changing the outcome, the C-nCas9 method is utilised to add or remove gene cassettes that are 1 kb or less in Lactobacillus casei, because Cas9 was created to accurately objective 2 neighboring locations on diverse branches, it has been employed as a GE editing technique with enhanced focus by concentrating on extra DNA’ sequences and resulting in broken strands. The expansion of techniques for detecting target effects in mammalian cells remained significant challenges.
Recent methods for detect target impacts have been developed. In order to overcome this issue, work is currently done to create Cas destruction that can overcome the PAM pattern's limitation. One Cas9 alternative to facilitate preserve distinguish PAMs of diverse sequence, such as “NG, GAA, and GAT”, was developed using virus-assists evolution.
The orthodox analysis-created chimeric Cas9 and the framework-guided mutagenesis-produced Cas12a (Cpf1) variant also demonstrate the impacts of PAM domain expansion and Distance influence decrease. These methods may be used to introduce desired sequences of DNA’and RNA into the GE in order to build organisms with the necessary features.
Editing Single Nucleotides
GE modification at the single strand level is almost always effective even in creatures with a basic GE, despite the risk of broken double-strands happening even in the situation of a mistake within the altered objective DNA’and the guidance RNA. The development of base operator and prime editor, 2 CCGE editing methods, has made it possible to limit the target effect without causing break in both strands (Figure 1).
A base editor (BE) that inserts position mutation addicted to the target DNA’ lacking DSB is produced through combining dCpf1, dCas9, and nCas9 with an RNA enzyme. Using a newly residential “Glycosylase Base Editor (GBE)”, base transformations from A to C and G to C might be facilitated.
These three enzymes nCas9, cytosine deaminas, and uracil “DNA” glycosylase (Ung) combine to form this complex. It convert A to C with an standard restriction specify of 92,9 % in E. The GE may be altered in a number of ways, together with insertion, deletion, and amplification, depends on the guidelines provided by the pegRNA GE.
Target Cleavage and Detection
Every time the Cas9-sgRNA complex recognizes a certain DNA’ strand as a target, a double-strand target DNA’ is preserved and may be recognized in a number of ways. For instance, a traction switch strategy has been anticipated to use PAM sequences to construct strain variant sequences in order to identifies Zika virus strains using C-Cas9.
If there is a difference in the viral DNA’ strand that precludes the Cas9/sgRNA complex from cleaving the objective DNA, the completely trigger mRNA is then generated for activating the firm grip switch. Likewise the coronavirus 2 SARS single nucleotide variant recognition assay called FELUDA was developed. The nucleolytic activity of the RuvC and HNH enzymatic regions of Cas9 has been abolished with the expansion of disabled Cas9 (dCas9). There has been work on developing a test to see whether the dCas9/sgRNA complexes have bonded to the target DNA.
Nucleic Acid Diagnostics
A diagnostic biomarker may be created utilizing the nucleic acid sequence that distinguishes each disease-causing viral or microbial infection. The CC complex enables the detection and cutting of a desired DNA’ by CC-Cas-based DNA’ editing tools.
Cas9 destruction cause breaks in two strands in a target DNA’ strand. By use of indels and consequent different end joining, the target gene is made inactive. A new DNA’ strand that is balancing to the pegRNA succession on the nick filament is polymerized by the RT. the attendance of a specific sequence of nucleics acid in the sample (Figure 2).
TransCleavage Activity
To make the Cas protein’s significantly nuclease function active, the gRNA and Cas mixture binds to the target nucleic acid. Trans-cleavage is a generic breakage behavior on ssDNA’in the region that Type V and Type VI Cas nucleases exhibit after cleaving the desired DNA/RNA in an arrangement with an illustrative RNA.
The objective cleavage, nevertheless, renders their nuclease activity inactive. Based on this occurrence, a diagnostic tool have been developed that allows for the indirect detection of Cas12 or Cas13 trans-cleavage activity utilizing techniques like fluorescence, color change potential difference, or others.
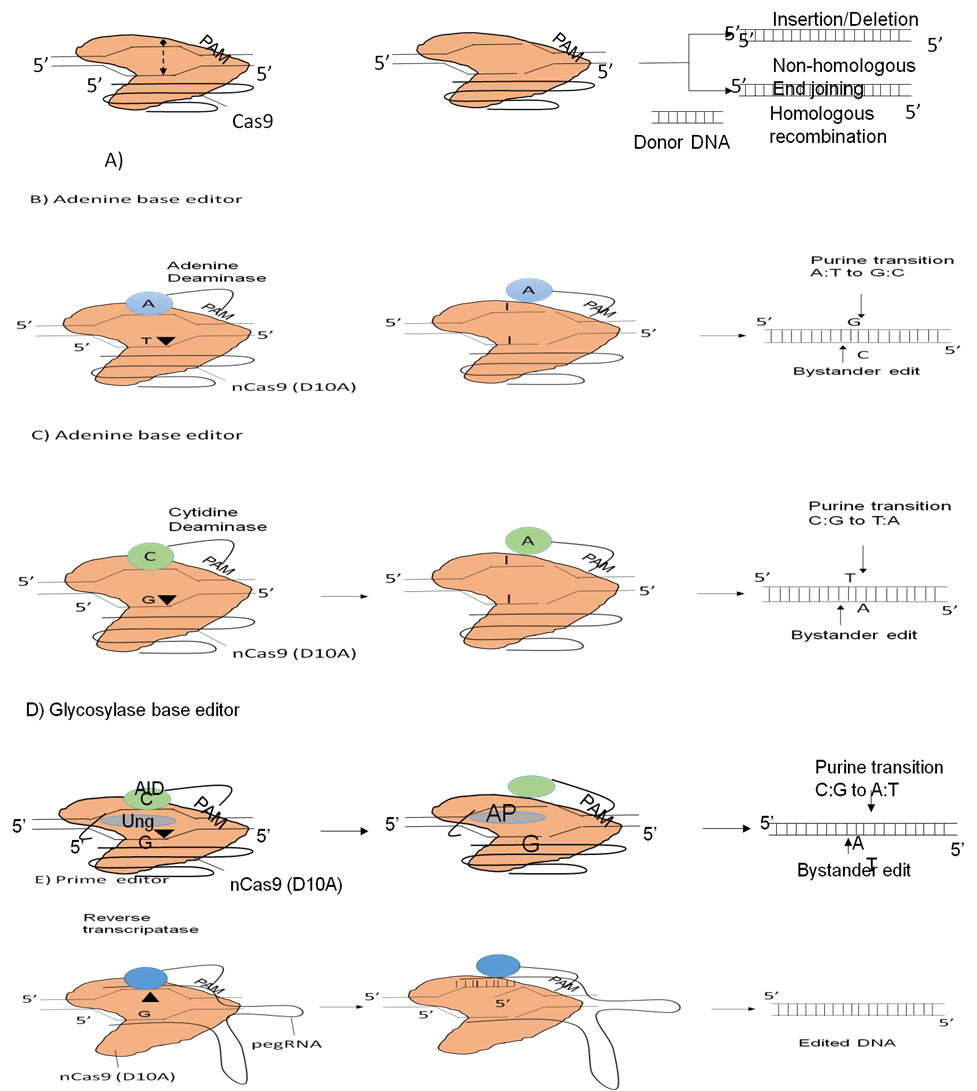
Figure 1. Genome editing instruments based on CC-Cas
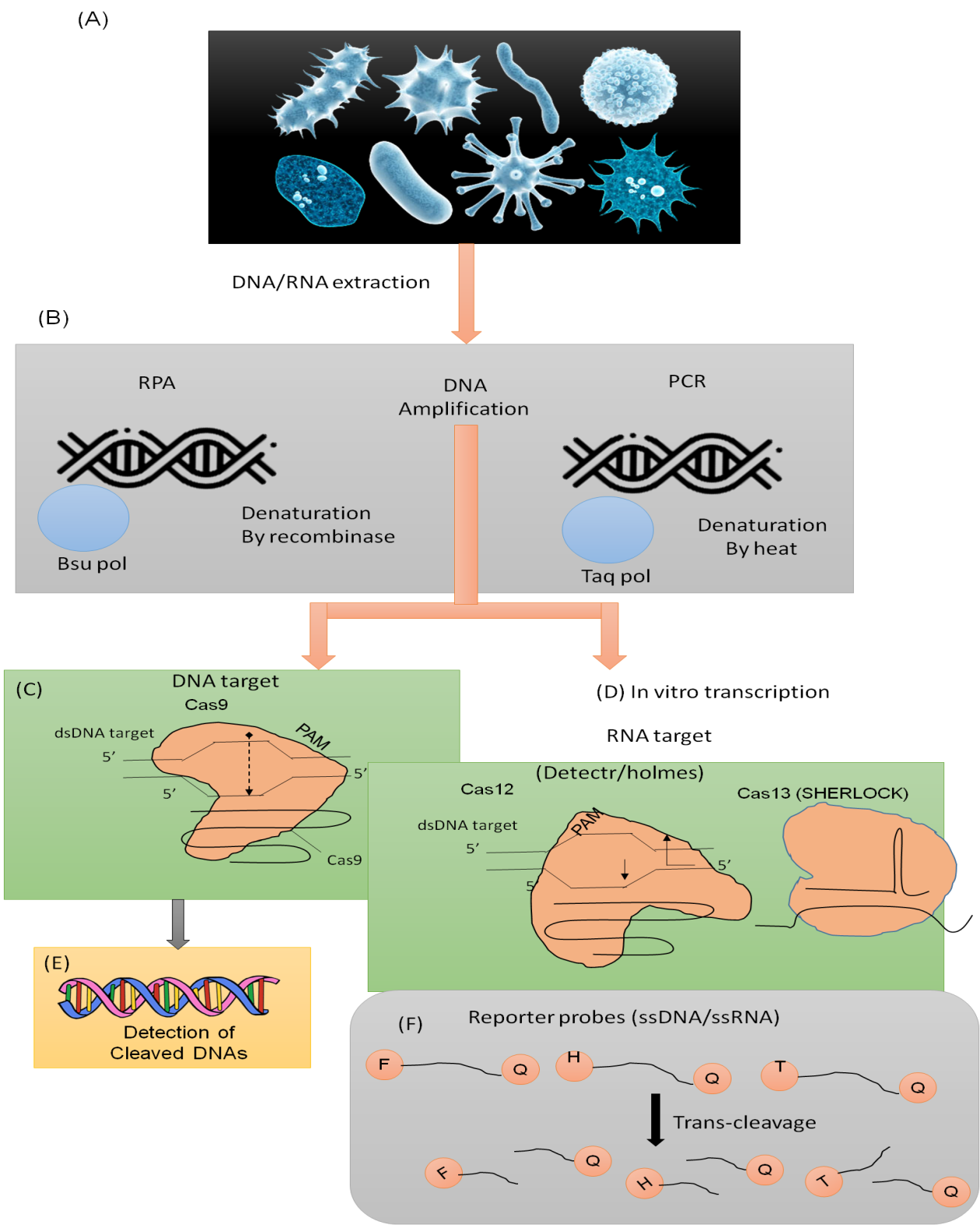
Figure 2. CC-Cas-based DNA’analysis
Cas12 has a PAM sequence that is predominantly located at the 5 end of the target, in contrast to Cas9, which recognize dsDNA’and ssDNA’as target and has a T-rich ‘PAM’ sequence at the 3 end of the intention. It may also reveal a particular nucleotide polymorphism (SNP) position in the objective DNA’ of the individual GE.
Using this method, the ‘SARS-CoV-2’ variant and African swine fever viruses are found. In contrast to HOLMES, the DNA’ Endonuclease-Target CC Trans Reporter (DETECTR) is an indicative method that can differentiate between different strains of ‘HPV’.
This is accomplished by first isothermally amplifying target DNA’ in the model among ‘RPA’ and then detects the fluorescent indicator of the ‘ssDNA-FQ’ journalist generate by trans-cleavage following the cleavage of the ‘dsDNA’ target of Cas12a. This method has been used to identify ‘SNP’s between Bacillus anthracis and B. cereus, which paves the way for the identification of particular species.
In addition, a variety of infectious agents, such as ‘MRSA’ and Mycoplasma pneumoniae, have been isolated. In a single reaction system, the All-In-One Dual CC-Cas12a (AIOD-CC) technology may be used to detect nucleic acids without the need to execute a separate phase of pre-amplification beforehand.
In vivo CC Regulation
Deactivated Cas9 (dCas9), a kind of ‘CC that developed into a tool for ‘GE editing in addition to controlling gene transcription. Fusion of different effector domain, such as transcripted activators or repressors, has allowed the CC-dCas system to be extended. By inquisitive with the necessary of RNA polymerase or transcripts factors via a process known as CC interference, the binding of the sgRNA-dCas protein multipart to the target DNA’ may prevent the start and elongated of transcripted, as illustrated in figure 3.
|
Table 2. CC-mediated gene control |
|||||
|
Efector domain |
Organism |
Target |
dCas protein |
Feature |
Species |
|
None |
Bacteria |
mRFP, sfGFP |
dCas9 |
Silencing effectiveness considerations |
Escherichia coli |
|
ω sub unit |
|
prsA, nprB, bpr, vpr |
dCas9−activator |
OAPS promoter modification increases BLA production 260-fold. |
Bacillus subtilis |
|
nothing |
|
pck, hom, pgi, gltA |
dCpf1 |
One crRNA array represses four target genes. |
C.glutamicum |
|
p300 |
Fungi |
breF, fuml, fwnA |
dCas9−activator |
Histone modification by dCas9 regulates tertiary metabolic genes. |
Aspergillus niger |
|
Mxi1 |
Yeast |
ku70, ku80 |
dCas9 |
Enhancing repression with Mxi1-dCas9 to increase HR rates |
Yarrowia lipolytica |
|
VP64 |
|
GFP |
dCas9−activator |
CCTFs may be activated and repressed by targeting different locations. |
S. cerevisiae
|
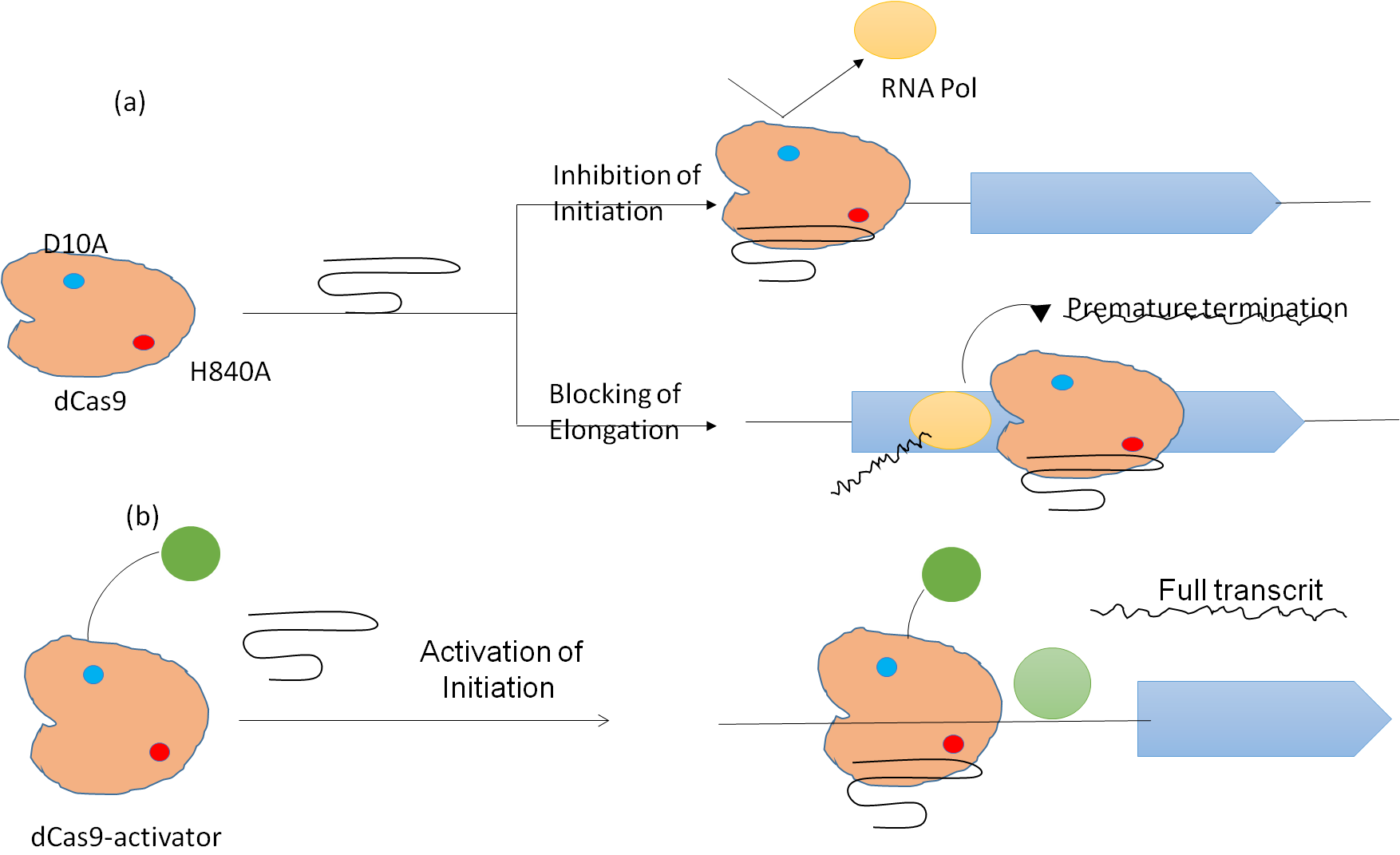
Figure 3. Deactivated Cas proteins regulate transcription
Multiple target genes may have their transcription reversibly inhibited by the CCi technology. The development of CC activation (CCa), which encourages the employment of transcription activators to target DNA’sequences, may trigger gene expression.
In a bacterial system, dCas9 has been coupled with a number of transcriptional activators, including as the RNA polymerases subunit and the phage activator Asia. When the sgRNA is positioned upstream of the TATA box, dCas9 coupled with VP64 significantly increases the movement of a journalist gene in S. Table 2 lists representative research on CC-mediate microbial gene transcription regulation.
Cellular System Optimization
Although TFs typically have a considerable energetic range, their orthogonality, modularity, and programmable are constrained, making them less suitable for synthetic biology. CC-based DNA’circuits may be built to quickly intention and control specific genes in intricate regulatory networks inside cells.
Logic circuits like AND, NOR, and NIMPLY are among the most common CC-based synthetic circuits in prokaryotes. A cellular sensor network may be connected to the CCi/a circuit to enable it to regulate host metabolism in response to environmental cues. A number of gRNAs must share a small intra-cellular supply of dCas9 in the casing of complicated gene circuits, which might lessen target gene suppression.
To address this issue, a non-toxic form of dCas9R1335K with reduced PAM recognition was created and combined with a PhlF inhibitor. As a result, the ‘dCas9-based circuit propose has been advanced in metabolic engineers and synthetic biology. CC-mediated gene control technology has been used to pinpoint chemical-genetic interactions and improve the biosynthetic pathways of different metabolites in microorganisms.
Target genes, even crucial ones, have mostly been inhibited using CCi, and carbon fux has been directed toward a desired good or bioactive substance. Additionally, to optimize the synthesis of desired metabolites, CC-mediated gene circuits or biosensors are employs to alternatively switch between cell growth and production phase. Additionally, by integrating ‘CC i/a technology with other metabolic engineering methods like deletion, overexpression of certain genes, or growth medium modification, metabolite production may be enhanced even more.
CONCLUSION
Several species, including prokaryotes and human cells, have used CC technology as a GE-edited tool. The precise GE editing and diagnostics, respectively, CC is employed for its highly specific and trans-cleaving nucleolytic activity. In addition to controlling the transcription of target genes, deactivated Cas protein is also employed in CC screening to identify the function of genes linked to certain disorders. In the future, codon optimization and well-established genetic vectors will be used to apply CC-based GE-edited techniques to varieties of cells, from bacteria to humans.
Customized strain creation will alter the soil and gut microbiome, which will enhance agricultural productivity and quality and allow for the control of individual health. Additionally, CC will be used to create a rapid and simple biosensor that can quickly and easily identify genetic markers and harmful microbes without the need for costly equipment, which will be very beneficial for diagnosing and treating diseases.
By emphasizing benefits including simplicity of use and scalable of modulated components, CC equipment has emerged as a fundamental tool in the fields of microbial metabolic engineering and synthetics biology. To address pressing issues including healthcare, pandemics, food shortages, and environmental pollution, synthetic biology's design-build-test-learn cycle has been speed up. This will contribute to the realization of a sustainable future for humans.
REFERENCE
1. Shan L, Dai Z, Wang Q. Advances and opportunities of CC/Cas technology in bioengineering non-conventional yeasts. Front Bioeng Biotechnol. 2021; 9:765396.
2. Zhang F. Development of systems for GE editing and beyond. Q Rev Biophys. 2019;52:e6.
3. Liu W, An C, Shu X, Meng X, Yao Y, Zhang J, Chen F, Xiang H, Yang S, Gao X, Gao SS. A dual-plasmid CC/Cas system for mycotoxin elimination in prokaryotic industrial fungi. ACS Synth Biol. 2020;9(8):2087-2095.
4. Nidhi S, Anand U, Oleksak P, Tripathi P, Lal JA, Thomas G, Kuca K, Tripathi V. Novel CC–Cas systems: an updated review of the current achievements, applications, and future research perspectives. Int J Mol Sci. 2021;22(7):3327.
5. Shin J, Yoon T, Park J, Park KS. Sensitive and simultaneous detection of hygiene indicator bacteria using an enhanced CC/Cas system in combination with a portable fluorescence detector. Sens Actuators B Chem. 2022;365:131871.
6. Ding W, Zhang Y, Shi S. Development and application of CC/Cas in microbial biotechnology. Front Bioeng Biotechnol. 2020;8:711.
7. Singh R, Chandel S, Ghosh A, Dey D, Chakravarti R, Roy S, Ravichandran V, Ghosh D. Application of CC/Cas system in the metabolic engineering of small molecules. Mol Biotechnol. 2021;63:459-476.
8. Mondal S, Halder SK, Mondal KC. Tailoring in fungi for next-generation cellulase production with special reference to CC/CAS system. Syst Microbiol Biomanuf. 2022;1-17.
9. Zheng Y, Li J, Wang B, Han J, Hao Y, Wang S, Ma X, Yang S, Ma L, Yi L, Peng W. Endogenous type I CC-Cas: from foreign DNA defense to prokaryotic engineering. Front Bioeng Biotechnol. 2020;8:62.
10. Liu Z, Dong H, Cui Y, Cong L, Zhang D. Application of different types of CC/Cas-based systems in bacteria. Microb Cell Fact. 2020;19(1):1-14.
11. Goh YJ, Barrangou R. Harnessing CC systems for precision engineering of designer probiotic lactobacilli. Curr Opin Biotechnol. 2019;56:163-171.
12. Barros JHT, de Carvalho Oliveira L, Cristianini M, Steel CJ. Non-thermal emerging technologies as alternatives to chemical additives to improve the quality of wheat flour for breadmaking: a review. Crit Rev Food Sci Nutr. 2023;63(11):1612-1628.
13. Roointan A, Morowvat MH. Road to the future of systems biotechnology: CC-Cas-mediated metabolic engineering for recombinant protein production. Biotechnol Genet Eng Rev. 2016;32(1-2):74-91.
14. Qiu M, Zhang J, Pang L, Zhang Y, Zhao Q, Jiang Y, Yang X, Man C. Recent advances in CC/Cas system-enabled portable detection devices for on-site agri-food safety assay. Trends Food Sci Technol. 2022.
15. Singh R, Chandel S, Ghosh A, Dey D, Chakravarti R, Roy S, Ravichandran V, Ghosh D. Application of CC/Cas system in the metabolic engineering of small molecules. Mol Biotechnol. 2021;63:459-476.
16. Mondal S, Halder SK, Mondal KC. Tailoring in fungi for next-generation cellulase production with special reference to CC/CAS system. Syst Microbiol Biomanuf. 2022;1-17.
17. Goh YJ, Barrangou R. Harnessing CC systems for precision engineering of designer probiotic lactobacilli. Curr Opin Biotechnol. 2019;56:163-171.
18. Yu HY, Wang SG, Xia PF. Reprogramming microbial CO2-metabolizing chassis with CC systems. Front Bioeng Biotechnol. 2022;10.
19. Fontana J, Voje WE, Zalatan JG, Carothers JM. Prospects for engineering dynamic CC–Cas transcriptional circuits to improve bioproduction. J Ind Microbiol Biotechnol. 2018;45(7):481-490.
20. Zheng Y, Han J, Wang B, Hu X, Li R, Shen W, Ma X, Ma L, Yi L, Yang S, Peng W. Characterization and repurposing of the endogenous Type IF CC–Cas system of Zymomonas mobilis for GE engineering. Nucleic Acids Res. 2019;47(21):11461-11475.
21. Jeong SH, Lee HJ, Lee SJ. Recent Advances in CCTechnologies for Synthetic Biology. J Microbiol. 2023;1-24.
22. Qiu M, Zhang J, Pang L, Zhang Y, Zhao Q, Jiang Y, Yang X, Man C. Recent advances on CC/Cas system-enabled portable detection devices for on-site agri-food safety assay. Trends Food Sci Technol. 2022.
23. Guo M, Chen H, Dong S, Zhang Z, Luo H. CC gene editing technology and its application prospect in medicinal plants. Chin Med. 2022;17(1):33.
24. Shmakova AA, Shmakova OP, Karpukhina AA, Vassetzky YS. CC/Cas: History and Perspectives. Russ J Dev Biol. 2022;53(4):272-282.
25. King RB, Sheldon JK, Long GM. Practical environmental bioremediation: the field guide. CRC Press; 2023.
FINANCING
No financing.
CONFLICT OF INTEREST
None.
AUTHORSHIP CONTRIBUTION
Conceptualization: Umar Farooq, Malathi Hanumanthayya, Izharul Haq.
Research: Umar Farooq, Malathi Hanumanthayya, Izharul Haq.
Writing - proofreading and editing: Umar Farooq, Malathi Hanumanthayya, Izharul Haq.