REVIEW
The Possibilities of Gene Drives for Managing Populations and Controlling Diseases
Posibilidades de los impulsores genéticos para la gestión de poblaciones y el control de enfermedades
Vijay Upadhye J1 ![]() *, Uzma Noor Shah2
*, Uzma Noor Shah2 ![]() *,
Basavaraj Mudhol3
*,
Basavaraj Mudhol3 ![]() *
*
1Department of Microbiology, Parul Institute of Applied Sciences. PO Limda, Gujarat, India.
2Department of Genetics, School of Sciences, JAIN (Deemed-to-be University). Karnataka, India.
3College of Nursing, Teerthanker Mahaveer University. Uttar Pradesh, India.
Cite as: Upadhye J V, Noor Shah U, Mudhol B. The Possibilities of Gene Drives for Managing Populations and Controlling Diseases. Salud, Ciencia y Tecnología. 2023;3(S1):451. https://doi.org/10.56294/saludcyt2023451
Submitted: 01-06-2023 Revised: 24-06-2023 Accepted: 01-08-2023 Published: 02-08-2023
Editor: Dr.
William Castillo-González ![]()
Associate Editor: Fasi
Ahamad Shaik ![]()
ABSTRACT
The technical limitations and the use of gene drives to address ecological problems by modifying all populations of wild species remain primarily speculative. Here, we examine the possibility that RNA-guided gene drives based on the CRISPR nuclease Cas9 could be used as an all-encompassing approach for introducing changed features into natural populations over a long period. We outline potential capabilities and possible disadvantages and offer new preventative measures to stem from the propagation of genes and undo genetic modifications. Editing the sexual animal population would significantly benefit both people and the environment. For instance, RNA-guided gene drives may stop the spread of illness, assist farming by correcting bug and weed chemicals and resistance to herbicides, and manage harmful invasive species. However, each prospective use needs to be carefully evaluated due to the likelihood of unfavorable ecological repercussions and the near inevitability of dissemination beyond political boundaries. To investigate the responsible application of this now hypothetical technology, we want serious, inclusive, educated public conversations.
Keywords: Diseases; Gene; Drives; Deoxyribonucleic Acid (DNA); Ribonucleic Acid (RNA); Agriculture.
RESUMEN
Las limitaciones técnicas y el uso de impulsores genéticos para abordar problemas ecológicos modificando todas las poblaciones de especies salvajes siguen siendo principalmente especulativos. Aquí examinamos la posibilidad de que los impulsores genéticos guiados por ARN y basados en la nucleasa CRISPR Cas9 puedan utilizarse como un enfoque global para introducir características modificadas en poblaciones naturales durante un largo periodo. Esbozamos las capacidades potenciales y las posibles desventajas y ofrecemos nuevas medidas preventivas para detener la propagación de genes y deshacer las modificaciones genéticas. La edición de la población animal sexual beneficiaría notablemente tanto a las personas como al medio ambiente. Por ejemplo, los impulsores genéticos guiados por ARN podrían detener la propagación de enfermedades, ayudar a la agricultura corrigiendo los bichos y malas hierbas químicos y la resistencia a los herbicidas, y gestionar especies invasoras dañinas. Sin embargo, cada uso prospectivo debe evaluarse cuidadosamente debido a la probabilidad de repercusiones ecológicas desfavorables y a la casi inevitabilidad de la diseminación más allá de las fronteras políticas. Para investigar la aplicación responsable de esta tecnología ahora hipotética, queremos conversaciones públicas serias, inclusivas y educadas.
Palabras clave: Enfermedades; Genes; Drives; Ácido Desoxirribonucleico (ADN); Ácido Ribonucleico (ARN); Agricultura.
INTRODUCTION
Despite significant advancements, the science of genetics has frequently struggled to solve critical biological issues that impact the planet and the general population. It has been challenging to modify the genetic code of essential hypothetical organisms until recently. Additionally, modified features often result in lower evolutionary fitness and are subsequently eradicated by natural selection. This limitation has significantly reduced their capacity to change environments through biological processes.(1) Those qualities might propagate to most individuals in indigenous populations over some time if we could devise a broad mechanism to ensure that artificial genes are instead favored by natural selection. With this power, we might tackle many pressing global issues, such as the spread of illnesses caused by insects, the increase of herbicides and resistance to pesticides, and the harm that invading species drive to the planet and agriculture.(2) The feature article explores the potential for employing genes that target insects to stop the spread of malaria. The challenge of designing homing endonucleases to cleave novel sequences of interest has slowed down progress. Because the genes encoding them are recurring, efforts to construct Gene drives that used TALENs and nucleases with zinc-finger nucleases that could be more easily retargeted were unreliable.(3) They are now much better equipped to modify the genetic material of various animals thanks to the subsequent invention and creation of the RIBONUCLEIC ACID-guided CRISPR nuclease. CRISPR is a unique enzyme initially identified from the 'CRISPR' acquired defense systems of bacteria. By merely expressing 'guide RIBONUCLEIC ACID' the target “DEOXYRIBONUCLEIC ACID sequence”, CRISPR can be instructed to cut virtually any DEOXYRIBONUCLEIC ACID sequence. It has made it possible to insert, delete, and replace genes in many other species in just over a year from the initial presentations in cells in humans.(4) It makes sense to construct RIBONUCLEIC ACID-guided genetic drives using the nuclease to get around the target and stability issues preventing the creation of genetic enterprises. The degree to which the particular capabilities are well-suited to overcome additional biochemical and evolutionary hurdles unique to developing secure and beneficial genetic drives is less evident.(5) We argue that this will likely allow researchers to build effective RIBONUCLEIC ACID-guided gene-driven systems in various species, not just mosquitoes. This development would constitute a whole new method of ecological engineering with numerous potential uses critical to human health, agriculture, biodiversity, and environmental science, in addition to changing the number of insects to stop them from transmitting illnesses.
DEVELOPMENT
Related works
This study is consistent with the functions mentioned in the research that led to the technologically feasible classification. It is crucial to look into the typical impacts of resistance development or detrimental changes of these drives in experiments across multiple generations in the same targeted species, aside from this basic categorization.(6) They examined full-length SARS-CoV-2 genomes from several nations in this study to identify early tendencies in the evolutionary structure of the novel COVID-19 virus. As an effect of good pressure at particular accessory genes ORF3a and ORF8 residues, the findings showed that SARS-CoV-2 developed into at least three phylogenetic groupings very early in its evolutionary history. Furthermore, they identify putative essential sites that are the subject of favorable choice at specific locations where the non-structured proteins nsp6 and helicase are present. They propose four possibly significant residues that might have influenced the initial development of SARS-CoV-2 in human beings based on the outcomes of various evolutionary algorithms and supported by the number of sequences impacted by this variant.(7) They now provide the TARE drive (Toxin-Antidote Recessive Embryo). When an intended gene is disrupted, hereditary fatal alleles result, saving drive-carrying organisms with a recorded aim expression. Modeling demonstrates that such drive will exhibit threshold-dependent invading factors, only spread when injected above a fitness-dependent regularity. According to Their findings, TARE drives are viable possibilities for creating efficient, adaptable, and spatially constrained drives for population growth.(8) They make a modified gene-drive rescue mechanism for Anopheles stephensi. This malaria vector lessens the load on females by the drive's integration into the gene encoding kynurenine hydroxylase by restoring its function. Rare functioning resistance variants do not stop drive assault, but ineffective resistance genotypes are eliminated by a dominant mate Ribonucleic acid l impact and a slower-acting standard selection mechanism. As a result, in An. Stephens, the Reckh process transforms a population suppressing (nRec) into a productive population modifying system. In addition, melanogaster has provided proof-of-concept for CRISPR-based rescuing methods that use modified sequences, this work describes the use of such systems in mosquitoes.(9) This study shows that CRISPR off-target editing may be kept to a minimum in the setting of a mosquito's gene drive. This is a significant advancement in knowledge of and management of CRISPR specificity in mosquitoes. Their findings demonstrate that gambiae homing-based genetic drive can be adequately planned for no detectable genetic inaccurate alterations.(10) According to their studies, homing forces are more likely to select for resistance than toxin-antidote pathways among gene drives that produce a mild reduction. Designed resistance appears to be a clear-cut safeguard against unintentional escaping, and it unquestionably provides a solid line of defense against highly destructive forces.(11) They investigate the haplodiploids' homing suppression gene drives and discover that a drive aimed at a female reproductive gene may still be effective. According to Their findings, this is feasible in haplodiploid living things, and a highly effective drive can result in potent and effective suppression. This is still the case even in more challenging conditions with continuous space, where the 'chasing' phenomena might impede or prohibit suppressing.(12)
Biological gene drives
In the wild, some genes 'drive' their own across communities by raising the likelihood that they will be transmitted. Transposons are viruses that place copies of their own into other parts of the genome. These endonuclease genes copy themselves into chromosomal lacking them, division distorters that wipe out rival chromosomal during meiosis, Medea components that reduce rival cousins who do not inherit them, and inheritable microbes like Wolbachia are examples of this natural gene drive.
Endonuclease gene drives
The drive is demonstrated by naturally migrating endonuclease genes by removing the relevant region of the chromosome that lacks them. By replicating the nuclease genes onto the broken chromosome by homologous recombination (HR), the cell is prompted to fix the break (figure 1a). The endonuclease-containing cassettes used for cloning are frequently described as a "gene drive" or just a "drive," while the cloning procedure is known as "homing." These gene can spread through a community even when they lower the reproductive fitness of the particular organisms that carry them because cloning increases the percentage of individuals that It's a tape the recording to be larger than 1/2 (figure 1b).(13) This self-sufficient approach might conceivably for a gene drive to propagate from a few individuals to all people in a population over multiple generations.
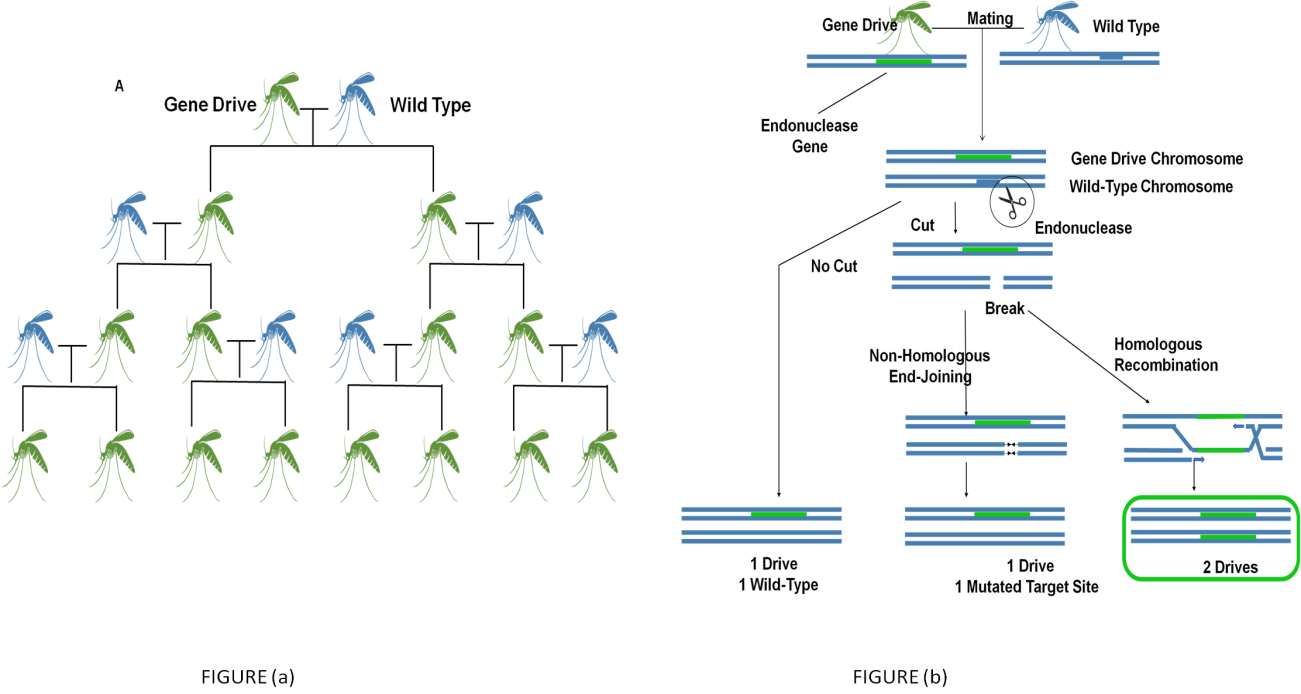
Figure 1. The spread of endonuclease gene drives
Endonuclease gene drives are prevalent. (A) All progeny preferentially inherit the gene drive when an natural harboring Pairing with a normal-form organism is the endonuclease gene impulse (blue). Even if the desire is just marginally harmful to the organism, this may allow it to increase until it affects every community member. (B) Due to the fact that the endonuclease destroys the associated wild-type genes, endonuclease gene drives are selectively transmitted. The gene drive chromosome must be used as a repair model for homology recombination, which involves replicating the campaign to the wild-type chromosome. Effective genetic drives must consistently cut when a repair directed by homology is most likely. Otherwise, the drive will not be duplicated if the endonuclease fails to cut or the cell uses another non-homologous end-joining repairing route.
Engineered gene drives
An endonuclease transgenic must be substituted for a natural sequence that can be cut to construct a viral gene driven. It will propagate across vulnerable ecosystems if it can successfully cut this pattern in creatures with one transgenic and one native location, consistently cause cells to replicate the transgene, and avoid being too expensive to the organisms. Genomic alterations, along with phenotypes, are disseminated throughout populations by standard mechanisms. They might be used to convey other transgenic mice or to mess with already-existing genes, according to Burt's original research. The germination cells that develop into eggs or sperm soon after fertilization can undergo the gene drive (figure 1a, 1b) duplicating stage (figure 2c), or it can only happen in these cells, keeping the vast majority of the organism's somatic tissues with just one instance of the drive (figure 2d). The number of people that are being suppressed is smaller.(24) Austin Burt described a sophisticated method using gene driving forces behind genes that only result in death or sterility when both versions are destroyed. When present, these 'genetic load' causes would spread quickly among slightly handicapped heterozygotes and ultimately contribute to a demographic collapse or perhaps extinction as a result of the accumulating burden of regressive genes. Another strategy would imitate genuine "gametic" or "meiotic" urges that tilt the sex ratio. The Y gene (or the same thing in other gender classification structures) would, in this model, store an enzyme called endonuclease that, during male meiosis, cuts and destroys the X chromosome, ensuring that the majority of viable sperm contains a Y chromosome.(15) A population collapse or extermination will occur as a result of the female population steadily declining. Only the pace of spread is influenced by the mate's motion, time of generation, and other traits of the people being targeted; whether an ordinary DEOXYRIBONUCLEIC ACID The rate at which the desire will spread throughout an intended group depends on genetic elements like recruitment efficiency, fitness costs, and advancing stability.(16) Simulations, however, indicate that they are more susceptible to population-specific environmental factors and density-dependent selection because of the negative and complex impacts on gene load and sex biassing suppression drive. There hasn't been any research on designed endonuclease gene drives that may spread throughout a wild population. The Crisanti and Russell groups have created genetic purposes.
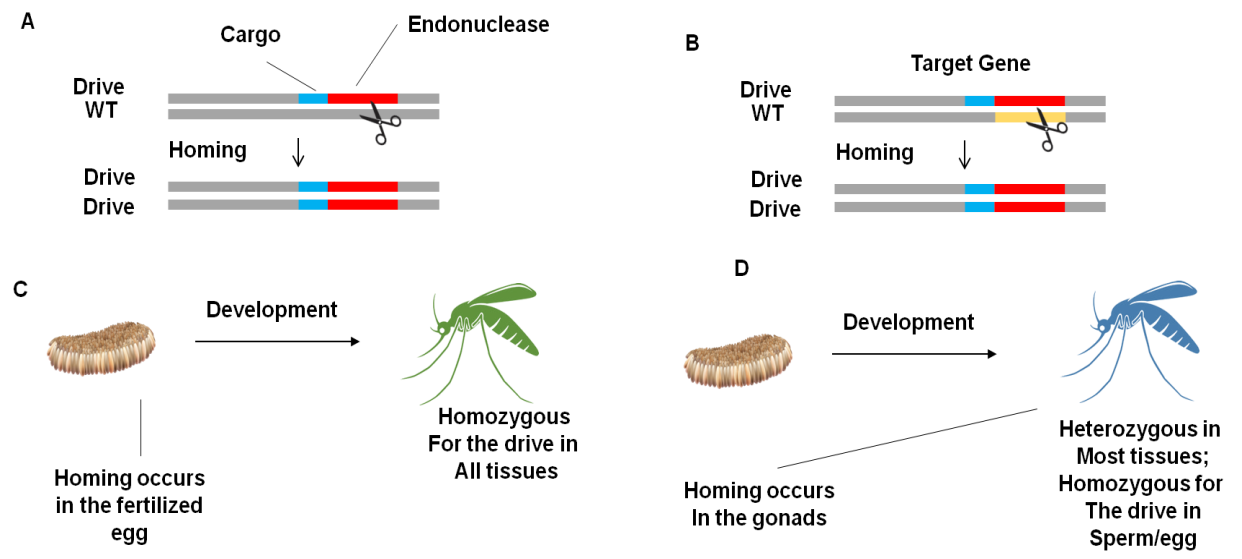
Figure 2. Gene drive propagation is governed by its effects and time. (A) Genetic drives are capable of transporting more genes. For instance, a transgenic that prevents malaria transmission may be spread across a population of wild mosquitoes. A cargo gene's ability to function is not maintained through selection. (B) DNA drives can alter or swap out other genes. For instance, a driver may swap out a mosquito gene essential to malaria. This tactic is stable during development since it can't propagate without affecting the target gene. (C) All organisms that possess drives will be heterozygous in all their organs if migration occurs in the zygote or early embryo. (D) The progeny will remain homozygous in most organs to prevent the effects of drive-induced disruptions if migration happens in the late germline cells that produce either eggs or sperm
Nevertheless, that can only propagate through the lab population of fruit flies and mosquitoes(17) genetically modified to contain the endonuclease break point. The gambiae Anopheles mosquito's X chromosome has a conserved region that repeats hundreds of times. A male-biasing suppressing drive is being developed by the Burt and Crisanti groups using an endonuclease that unintentionally cuts that sequence.(18) If practical, their efforts could significantly lower the number of this crucial malaria vector. The instinctual identification site of the appropriate enzyme is cut by all designed gene drives according to homing endonucleases. Despite initial optimism, it has been challenging to create homing endonucleases that cleave novel targeting sequences. Only a handful of labs have recently achieved this objective, despite numerous attempts over the past several years.(19) The fruit fly's gene drive has recently been improved by a team using TALENs or modular zinc-finger nucleases instead of focusing endonucleases,(20) both of which may be programmed to cut different targeting sequences. Due to the adaptive fragility of the module instances present in those protein molecules, both cutting and homing, while immediately efficient, lost some of their potency with a period.
Genome editing using RIBONUCLEIC ACID guidance using the CRISPR nuclease
A straightforward form of genome modification uses the identical endonuclease gene drive mechanism: it cuts the targeted gene. It provides the cell with a modified copy as a template for repairing the injury. TALENs and zinc-finger nucleases, both Adaptable proteins that can be changed or developed for targeting new patterns, were used for most eukaryotic genome modification projects over the past decade, though only in specialized labs.(21) The development of CRISPR, an protein that can be configured to cut target DEOXYRIBONUCLEIC ACID patterns stipulated by underlying RIBONUCLEIC ACID molecules, has made genome modification more accessible.(22) CRISPR is a part of bacteria's Type 2.
“CRISPR-acquired” immunity systems, which enable cells to "remember" sequences of previously seen viruses and defend themselves by recognizing and chopping those parts upon re-exposure. The way they do this is by putting DEOXYRIBONUCLEIC ACID segments into a stored position, transcribing to create RIBONUCLEIC ACIDs with the same process, and telling CRISPR to cut any matched sequences of DEOXYRIBONUCLEIC ACID.(23) Only segments flanking by a protospacer-adjacent motif (PAM) at the three ′ ends of the target "protospacer" sequence will CRISPR cut, and that's the only constraint. The most widely utilized CRISPR homolog may remove protospacers found in every eight base pairs since it only requires two bases for its PAM (NGG). Surprisingly, a single guide RIBONUCLEIC ACID (sgRIBONUCLEIC ACID) of less than 100 base pairings in sequence can instruct CRISPR to cut a particular protospacer in the genome.(24) This instruction RIBONUCLEIC ACID must start with a spacers segment made up of 17–20 base pairs that are equal to the desired gene. Selecting protospacers inside the desired gene, creating one or more guide RIBONUCLEIC ACIDs with compatible separators, and sending CRISPR, those guide RIBONUCLEIC ACIDs, and altered repaired templates devoid of those protospacers into a cell are the steps in the procedure for editing (Figure 3). A target gene is edited by selecting protospacers inside the gene, creating one or more RIBONUCLEIC ACID guides with corresponding separators, then sending CRISPR, those guide RIBONUCLEIC ACIDs, and altered repaired templates devoid of those protospacers into the nucleus (figure 3).
In one study, CRISPR is effective enough to reduce and alter several genes.(25) This enzyme is quite selective, removing only protospacers closely related to the spacers sequences of the guiding RIBONUCLEIC ACID, and it is active in a wide range of species. Furthermore, by selecting enlisting proteins that regulate linked to CRISPR or the guide RIBONUCLEIC ACID, techniques that allow CRISPR to bind but not cleave permit the regulation of the transcription of gene targets. These applications are all recent (2 years) creations. It is logical to wonder if it could be feasible to create genetic drives based on CRISPR because RIBONUCLEIC ACID-guided genome modification uses the same replicating process as endonuclease gene drives. Almost any genomic change produced using CRISPR may theoretically be disseminated throughout sexually reproduced organisms utilizing RIBONUCLEIC ACID-guided gene drives.
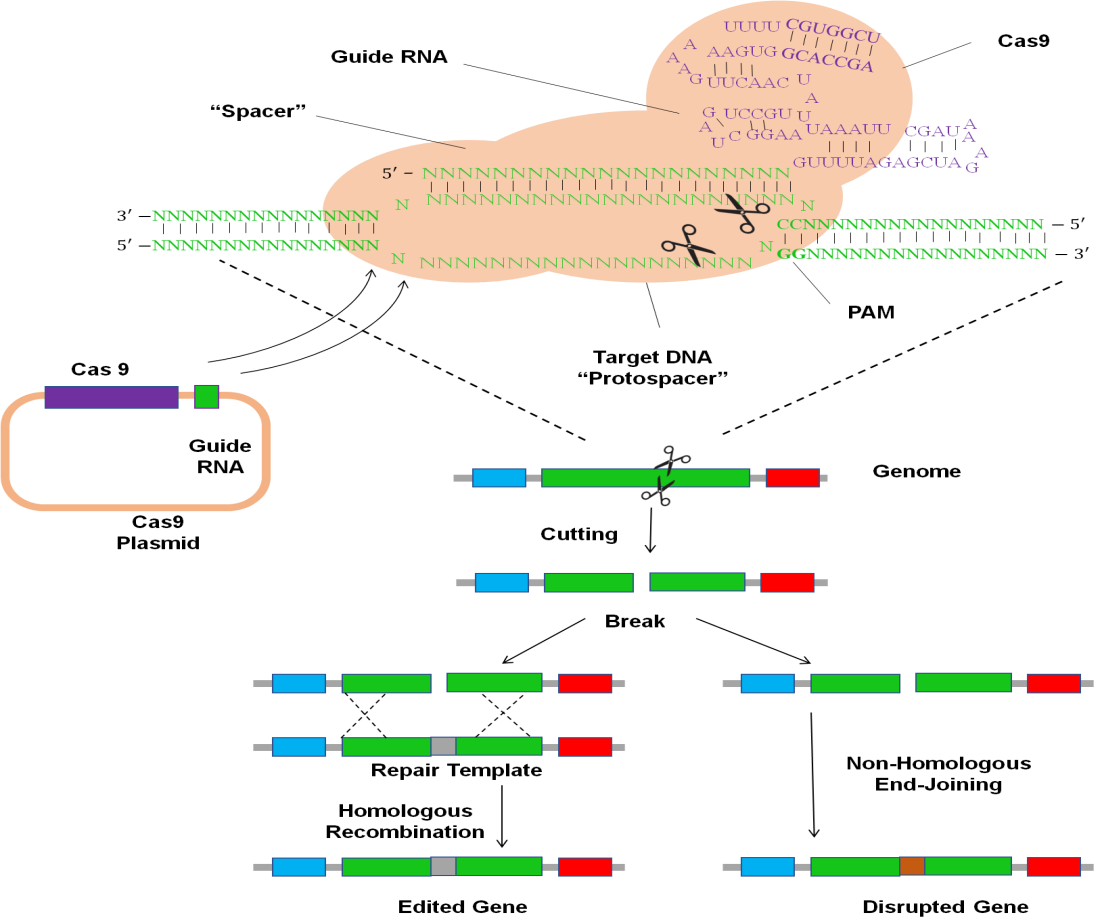
Figure 3. Cas9-based RNA-guided genome modification
Figure 3 shows Cas9-based RNA-guided genome modification. The target cell must first be introduced to the nuclease Cas9 protein and RNA that guides it. Although delivering RNA works just as well, transferring DNA production plasmids is frequently used. The guide RNA instructs Cas9 to attach to 'protospacer' regions in the desired DNA that coincide with the spacer region in the guide RNA. The protospacer-adjacent motif (PAM), which is NGG for the most widely used Cas9 protein, must be placed on either side of the protospacer. Cas9 will make blunt-ended double-strand breaks in the DNA if the separator and protospacer are identical or have a few mismatches at the 5′ ends of the spacer. If given a repaired template with the required modifications and similarity to the regions on By inserting the repair templates into the chromosome on Germline that produces sperm and eggs, the cell can use identical recombination to close the gap. If not, the break will be fixed by nonhomologous end-joining, damaging the gene. When editing numerous genes at once or both chromosomes simultaneously, Cas9 cutting is effective enough to do so. Future generations may inherit the alterations if the altered cell Egg and sperm-producing germline germ.
Gene drive limitations
Given that biological drive systems may be widely accessible, It's going to important to get a complete recognising of their primary restrictions. It takes a long time for gene drives to spread throughout populations, which is the first and most crucial fact. To begin spreading the gene drive throughout the wild humanity, transgenic organisms containing the gene drive must be out into the wild where they can mate with species of the native kind. The total length of time required for the virus to infect everyone individuals, the variety of drive-carrying individuals released, the living thing's time to generation, the efficacy of homing, the impact of a movement on one's own fitness, the nature of combining up and gene flow in the community, and the usefulness of locating all have an impact on how long it takes to propagate. A few centuries will pass.(26) In creatures that can reproduce quickly, drives will emerge quickly; yet, in species with long lifespans, drives will emerge gradually. Second, gene drive has little impact on creatures exclusively reproduced sexually by self-fertilization or clone divisions. This category includes every kind of bacteria, viruses, and most single-cell creatures. Some plants are examples of societies that use a combination of sexual and asexual reproduction, where highly effective characteristic motions may expand slowly. To avoid persecution, drives intended for controlling populations would probably be forced by compelling the target species to multiply usually.
Third, during an evolution period, genomic changes caused by drives are not irreversible. Genetic drives can propagate traits through a population even when expensive to each organism.(27) Still, once the purpose has reached obsession, damaging features will ultimately be surpassed by more fit variants. Extremely harmful elements may be eliminated even faster, with ineffective forms already being present in significant numbers before the drive and its shipment have a chance to affect the entire population. Once the drive the mechanism succeeds fixation, the strain of selection favouring the continuing function of CRISPR and the guide RIBONUCLEIC ACIDs will relax, even if the trait is entirely related to the drive mechanism. planned debuts of new RIBONUCLEIC ACID-guided gene drives will likely be necessary to continuously replace the damaged copies in the environment if harmful features are to be maintained within a population for an extended period. Fourth, research on mosquito-borne diseases has contributed significantly to understanding risk management and containment concerns related to genetic drives. Comparable methods assess the ecological impacts on mosquitoes and the few other animals for whom alternative genetic and biological control strategies have been studied. While these examples offer an essential point of departure for research into RIBONUCLEIC ACID-guided gene drives in different target animals, additional studies focusing on the specific drive, population, and surrounding ecology will be required.
Applications for gene drives directed by RIBONUCLEIC ACID
Figure 4 Preventing declines in population. A major may result from already hypothesized gene load and meiotic suppressive processes spreading unrestrictedly. To provide greater accuracy over the degree of repression, Other gene drive types could be used. Aside from increasing vulnerability to a specific chemical, which might therefore be used as a population-specific chemical, “sensitization drives” would be innocuous. The average frequency of drive copying occurrences and, hence, the degree of population suppression would be constrained in the presence of an evolutionarily ‘unstable drive.’ Only when ‘interacting drives’ come across a particular genetic signature in the community, in this example, a distinct gene drive, will suppression start. A sterile-daughter effect would result from the combination (Figure 4—figure supplement 1), Which could endure for several generations A vaccine comes last.
DNA drives that are guided by RIBONUCLEIC ACID have a chance to combine ecological and genetic control. They could help us address issues in worldwide health, farming, long-term viability research on the environment, and many other fields. Among these possibilities, limiting invasive plant populations that are harmful to the environment and the economy, managing agricultural pests, and decreasing the spread of vector-borne diseases may be the most convincing.(28)

Figure 4. Controlling population suppression
Eradicating insect-borne diseases
Infectious conditions carried by insects have a startling human toll. More than 650 000 people die yearly from malaria alone, most children, and 200 million more suffer from incapacitating fevers.(29) Bugs can also transmit Lyme disease, trypanosomiasis, yellow fever, chikungunya, trypanosomiasis, and leishmaniasis. By modifying vector species to obstruct transmission, many conditions may be managed or even wiped off. Candidate gene defects or transgenes that hinder the transmission of malaria and other well-researched disorders have been found in several laboratories.(30) These modifications may or may not enable the eradication of the disease before the pathogen develops opposition based on its efficacy. Alternatively, the pertinent species of vector may be inhibited or eradicated using RIBONUCLEIC ACID-guided DNA drives. Then when the illness has been stopped, they could be reintroduced from protected additional using RIBONUCLEIC ACID-guided gene transfers, the pertinent vector organisms may be suppressed or eliminated. They could then be reintroduced from secured laboratories or island populations once the sickness has been stopped. Gene drive tactics may be particularly useful in combating the issue of vectors of mosquitoes that have developed a predilection for striking and sitting outside in the context of malaria. These traits make them immune to the present control methods based on bednets and indoor insecticide spritzing.
Sustainable farming practices and safety
The development of insecticides and resistance to herbicides is a significant issue for farming. Resistance populations are expected to continue to be durable unless the pertinent genes have a substantial impact on fitness in the absence of pesticides or herbicides. This suggests that to restore fragility, RIBONUCLEIC ACID-guided sensitizing drives could swap out resistance genes for their ancestral equivalent. For instance, sensitizing goals could possibly reverse changes that render horseweed and the pigweed plant immune to glyphosate herbicides, which is important for long-term no-till farming, or allow the western corn rootworm to withstand Bt toxins.(31) Since these three species only produce a single generation each year, it is necessary to release a greater number of drive-bearing organisms for it to have an immediate effect. However, fewer than are currently published for controlling pests using the sterile-insect methodology. Removing would need to take place in nearby, non-pesticide or non-herbicide-treated regions, which would fast turn into repositories of sensitizing drives that may spread into neighboring fields.(32) Any specific pesticide or herbicides may be used indefinitely by periodically releasing new campaigns. It will be necessary to conduct modeling studies to assess the viability of various species of interest.
Potentially, a second type of sensitizing drive could make populations of pests susceptible to chemicals that have never before afflicted them. For instance, a gene crucial to fitness may be switched out for a variant from a different species or lab isolation whose function is responsive to a specific substance.(33) The ultimate creation and use of more secure and more particular to a species herbicides and insecticides might theoretically result from this method.
CONCLUSION
RIBONUCLEIC ACID-guided gene drives could have far-reaching effects, which calls for a careful and measured reaction. Before gene drives can be used for any of the above uses, numerous technical challenges must be addressed. Due to organic systems' complexity and engineering challenges, many of their hypotheses and forecasts will likely be wrong. The geneticists could quickly be able to edit the genetic code of living populations, undo or modify these changes in response to field information, and even carry out selective population repression, given the speed at which CRISPR research is currently progressing and the wide range of outcomes that can be gathered with even the most basic gene impulses. What criteria may apply to evaluate a RIBONUCLEIC ACID-guided gene engine designed to address a particular problem? Reducing human diseases transmitted by insects, creating and promoting more sustainable farming methods, and controlling invasive species that affect the natural world are all admirable objectives. However, there are legitimate worries about their capacity to foresee the effects of these actions on the environment and people. They intend to start open, inclusively, and well-informed dialogues about the ethical assessment and use of these emerging technologies by presenting these opportunities to the attention of the scientific community and the general public before they are realized in laboratories.
REFERENCES
1. Presley TD, Harp NA, Holt LS, Samuel D, Harp JJ. Incorporating physics principles in general biology to promote integrative learning and thinking. Int J Innov Teach Learn High Educ (IJITLHE). 2021;2(1):1-19.
2. Gause GF. The struggle for existence: a classic of mathematical biology and ecology. Courier Dover Publications; 2019.
3. Dhole S, Vella MR, Lloyd AL, Gould F. Invasion and migration of spatially self‐limiting gene drive: A comparative analysis. Evol Appl. 2018;11(5):794-808.
4. Sandler R. The ethics of genetic engineering and gene drives in conservation. Conserv Biol. 2020;34(2):378-385.
5. Bier E. Gene drives, gaining speed. Nat Rev Genet. 2022;23(1):5-22.
6. Wang GH, Du J, Chu CY, Madhav M, Hughes GL, Champer J. Symbionts and gene drive: two strategies to combat vector-borne disease. Trends Genet. 2022.
7. Cook F, Bull JJ, Gomulkiewicz R. Gene drive escape from resistance depends on mechanism and ecology. Evol Appl. 2022;15(5):721-734.
8. Dressler L, Bortolomeazzi M, Keddar MR, Misetic H, Sartini G, Acha-Sagredo A, Montorsi L, Wijewardhane N, Repana D, Nulsen J, Goldman J. Comparative assessment of genes driving cancer and somatic evolution in non-cancer tissues: an update of the Network of Cancer Genes (NCG) resource. Genome Biol. 2022;23(1):1-22.
9. De Graeff N, Jongsma KR, Lunshof JE, Bredenoord AL. They are governing gene drive technologies: a qualitative interview study. AJOB Empir Bioeth. 2022;13(2):107-124.
10. Hammond A, Karlsson X, Morianou I, Kyrou K, Beaghton A, Gribble M, Kranjc N, Galizi R, Burt A, Crisanti A, Nolan T. Regulating the expression of gene drives is key to increasing their invasive potential and mitigating resistance. PLoS Genet. 2021;17(1):e1009321.
11. Gomulkiewicz R, Thies ML, Bull JJ. Evading resistance to gene drives. Genetics. 2021;217(2):iyaa040.
12. Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. Evolutionary dynamics of CRISPR gene drives. Sci Adv. 2017;3(4):e1601964.
13. Campbell KJ, Saah JR, Brown PR, Godwin J, Howald G, Piaggio A, Thomas P, Tompkins DM, Threadgill D, Delborne J, Kanavy DM. A potential new tool for the toolbox: assessing gene drives for eradicating invasive rodent populations.
14. Wang GH, Du J, Chu CY, Madhav M, Hughes GL, Champer J. Symbionts and gene drive: two strategies to combat vector-borne disease. Trends Genet. 2022.
15. Metzloff M, Yang E, Dhole S, Clark AG, Messer PW, Champer J. Experimental demonstration of tethered gene drive systems for confined population modification or suppression. BMC Biol. 2022;20(1):1-13.
16. Yang E, Metzloff M, Langmüller AM, Xu X, Clark AG, Messer PW, Champer J. A homing suppression gene drive with multiplexed gRNAs maintains high drive conversion efficiency and avoids functional resistance alleles. G3. 2022;12(6):jkac081.
17. Devos Y, Mumford JD, Bonsall MB, Camargo AM, Firbank LG, Glandorf DC, Nogué F, Paraskevopoulos K, Wimmer EA. The potential use of gene-drive-modified insects against disease vectors, agricultural pests, and invasive species poses new challenges for risk assessment. Crit Rev Biotechnol. 2022;42(2):254-270.
18. Thumberger T, Tavhelidse-Suck T, Gutierrez-Triana JA, Cornean A, Medert R, Welz B, Freichel M, Wittbrodt J. Boosting targeted genome editing using the hei-tag. Elife. 2022;11:e70558.
19. Liu Z, Chen S, Xie W, Song Y, Li J, Lai L, Li Z. Versatile and efficient in vivo genome editing with compact Streptococcus pasteurianus CRISPR. Mol Ther. 2022;30(1):256-267.
20. Reichart D, Newby GA, Wakimoto H, Lun M, Gorham JM, Curran JJ, Raghuram A, DeLaughter DM, Conner DA, Marsiglia JD, Kohli S. Efficient in vivo genome editing prevents hypertrophic cardiomyopathy in mice. Nat Med. 2023;29(2):412-421.
21. Kim S, Jeong YK, Cho CS, Lee S, Sohn CH, Kim JH, Jeong Y, Jo DH, Bae S, Lee H. Enhancement of gene editing and base editing with therapeutic ribonucleoproteins through in vivo delivery based on absorptive silica nanoconstruct. Adv Healthc Mater. 2023;12(4):2201825.
22. Aliaga Goltsman DS, Alexander LM, Lin JL, Fregoso Ocampo R, Freeman B, Lamothe RC, Perez Rivas A, Temoche-Diaz MM, Chadha S, Nordenfelt N, Janson OP. Compact Cas9d and HEARO enzymes for genome editing were discovered in uncultivated microbes. Nat Commun. 2022;13(1):7602.
23. Dong C, Gou Y, Lian J. SgRNA engineering for improved genome editing and expanded functional assays. Curr Opin Biotechnol. 2022;75:102697.
24. Vento JM, Beisel CL. Genome editing with Cas9 in lactobacilli. In: Recombineering: Methods and Protocols. New York, NY: Springer US; 2022. p. 245-261.
25. Wang GH, Du J, Chu CY, Madhav M, Hughes GL, Champer J. Symbionts and gene drive: two strategies to combat vector-borne disease. Trends Genet. 2022.
26. Liu Y, Champer J. Modeling homing suppression gene drives in haplodiploid organisms. Proc R Soc B. 2022;289(1972):20220320.
27. Terradas G, Hermann A, James AA, McGinnis W, Bier E. High-resolution in situ analysis of Cas9 germline transcript distributions in gene-drive Anopheles mosquitoes. G3 (Bethesda). 2022;12(1):jkab369.
28. Xu X, Harvey-Samuel T, Siddiqui HA, Ang JXD, Anderson ME, Reitmayer CM, Lovett E, Leftwich PT, You M, Alphey L. Toward a CRISPR-Cas9-based gene drive in the diamondback moth Plutella xylostella. CRISPR J. 2022;5(2):224-236.
29. Bier E. Gene drives, gaining speed. Nat Rev Genet. 2022;23(1):5-22.
30. Koul B, Yakoob M, Shah MP. Agricultural waste management strategies for environmental sustainability. Environ Res. 2022;206:112285.
31. Bartolucci C, Scognamiglio V, Antonacci A, Fraceto LF. What makes nanotechnologies applied to agriculture green? Nano Today. 2022;43:101389.
32. An C, Sun C, Li N, Huang B, Jiang J, Shen Y, Wang C, Zhao X, Cui B, Wang C, Li X. Nanomaterials and nanotechnology for the delivery of agrochemicals: strategies towards sustainable agriculture. J Nanobiotechnology. 2022;20(1):1-19.
33. Liu Y, Li D, Du B, Shu L, Han G. Rethinking sustainable sensing in agricultural Internet of Things: From power supply perspective. IEEE Wireless Commun. 2022;29(4):102-109.
FUNDING
No financing.
CONFLICTS OF INTEREST
None.
AUTHOR CONTRIBUTIONS
Conceptualization: Vijay Upadhye J, Uzma Noor Shah, Basavaraj Mudhol.
Methodology: Vijay Upadhye J, Uzma Noor Shah, Basavaraj Mudhol.
Drafting - original draft: Vijay Upadhye J, Uzma Noor Shah, Basavaraj Mudhol.
Writing - proofreading and editing: Vijay Upadhye J, Uzma Noor Shah, Basavaraj Mudhol.