doi: 10.56294/saludcyt2024.1333
ORIGINAL
Relationship between genes pelA and biofilm density in clinical isolates Pseudomonas Aeruginosa
Relación entre los genes pelA y la densidad de biopelícula en aislados clínicos Pseudomonas Aeruginosa
Nanda Yaultsa1 ![]() , Puspa Wardhani2
, Puspa Wardhani2 ![]() *, Aryati2
*, Aryati2 ![]()
1Master of Basic Medical Sciences Student, Faculty of Medicine, Airlangga University, Surabaya, Indonesia.
2Department of Clinical Pathology, Faculty of Medicine, Airlangga University, Surabaya, Indonesia.
Cite as: Yaultsa N, Wardhani P, Aryati A. Relationship between genes pela and biofilm density in clinical isolates pseudomonas aeruginosa. Salud, Ciencia y Tecnología. 2024; 4:.1333. https://doi.org/10.56294/saludcyt2024.1333
Submitted: 21-02-2024 Revised: 19-07-2024 Accepted: 14-12-2024 Published: 15-12-2024
Editor: Prof.
Dr. William Castillo-González ![]()
Corresponding author: Puspa Wardhani *
ABSTRACT
Introduction: Pseudomonas aeruginosa is a Gram-negative pathogen that causes major infections in patients with weak body defense mechanisms. The pelA gene encodes the Pel polysaccharide for the production of cationic charged polysaccharides, and expression of the polysaccharide-encoding locus (pelA) by Pseudomonas aeruginosa is essential for biofilm formation.
Objective: analyzing the relationship between the pelA gene and biofilm density in clinical isolates of Pseudomonas aeruginosa.
Method: analytical observational research design with consecutive sampling, a total sample of 33 clinical isolates of Pseudomonas aeruginosa. The pelA gene was detected using a PCR tool (SimpliAmpTMThermal Cycler) and biofilm density examination was examined using a BioRad iMark Microplate Reader.
Results: thirty-three clinical isolates of Pseudomonas aeruginosa. In this study, the results of pelA gene detection showed that 31 (93,9 %) isolates were positive for the pelA gene and 2 (6,1 %) isolates were negative for the pelA gene. The results of the biofilm density examination showed that 4 (12,1 %) clinical isolates did not produce biofilm, 10 (30,3 %) clinical isolates had weak biofilm production, 13 (39,4 %) clinical isolates had moderate biofilm production, 6 (18,2 %) isolates had strong biofilm production. The pelA gene has a relationship with biofilm density (p = 0.011) with moderate relationship strength.
Conclusion: the pelA gene was moderately associated with biofilm density in clinical isolates of Pseudomonas aeruginosa. This suggests that the pelA gene plays an important role in biofilm formation in Pseudomonas aeruginosa strains which may contribute to its pathogenic properties, especially in patients with weakened immune mechanisms.
Keywords: Pseudomonas aeruginosa; Biofilm Density; PelA Gene.
RESUMEN
Introducción: Pseudomonas aeruginosa es un patógeno Gram-negativo que causa infecciones importantes en pacientes con mecanismos de defensa corporal débiles. El gen pelA codifica el polisacárido Pel para la producción de polisacáridos catiónicos cargados; la expresión del locus codificante de polisacáridos (pelA) por Pseudomonas aeruginosa es esencial para la formación de biopelículas.
Objetivo: analizar la relación entre el gen pelA y la densidad de biopelículas en aislados clínicos de Pseudomonas aeruginosa.
Método: diseño de investigación observacional analítico con muestreo consecutivo, muestra total de 33 aislados clínicos de Pseudomonas aeruginosa. el gen pelA se detectó utilizando una herramienta de PCR (SimpliAmpTMThermal Cycler) y el examen de la densidad de la biopelícula se examinó utilizando un lector de microplacas BioRad iMark.
Resultados: treinta y tres aislados clínicos de Pseudomonas aeruginosa. En este estudio, los resultados de la detección del gen pelA mostraron que 31 (93,9 %) aislados fueron positivos para el gen pelA y 2 (6,1 %) aislados fueron negativos para el gen pelA. Los resultados del examen de densidad de biopelículas mostraron que 4 (12,1 %) aislados clínicos no produjeron biopelículas, 10 (30,3 %) aislados clínicos tuvieron una producción débil de biopelículas, 13 (39,4 %) aislados clínicos tuvieron una producción moderada de biopelículas, 6 (18,2 %) Los aislados tuvieron una fuerte producción de biopelículas. El gen pelA tiene una relación con la densidad de la biopelícula (p = 0,011) con una fuerza de relación moderada.
Conclusión: el gen pelA se asoció moderadamente con la densidad de biopelículas en aislados clínicos de Pseudomonas aeruginosa. Esto sugiere que el gen pelA desempeña un papel importante en la formación de biopelículas en cepas de Pseudomonas aeruginosa, lo que puede contribuir a sus propiedades patógenas, especialmente en pacientes con mecanismos inmunitarios debilitados.
Palabras clave: Pseudomonas aeruginosa; Densidad de Biopelículas; Gen PelA.
INTRODUCTION
Pseudomonas aeruginosa is a gram-negative pathogen that causes infection in patients with a weakened immune system.(1) Bacteria Pseudomonas aeruginosa has been found in many cases of clinical infection in several parts of the body making treatment more difficult, The prevalence of Pseudomonas aeruginosa In Indonesia reached more than 30 %.(2) The highest case is Pseudomonas aeruginosa, which causes cystic fibrosis, respiratory tract infections (RTI), urinary tract infections (UTI), and wound infections. Infections can be found in many clinical samples, including sputum, urine, wounds, pus, feces, and post-surgical infections.(3) Pseudomonas aeruginosa can cause chronic infections, and bacteria in the body are often associated with the process of biofilm formation.(2,4)
Bacterial biofilms are complex microbial communities surrounded by extracellular polymeric substances. Biofilms are a significant problem in treating bacterial infections and are a major cause of infection persistence.(4) National Institutes of Health (NIH) revealed that among all microbial and chronic infections, 65 % and 80 % respectively are associated with biofilm formation. Biofilm formation is an important factor in the pathogenesis of Pseudomonas aeruginosa and isolates Pseudomonas aeruginosa biofilm producers are more resistant to antibiotics and immune responses.(4)
Pseudomonas aeruginosa produces at least three polysaccharides Alginate encoded by the AlgD gene, Pel encoded by the pelA gene, and Psl encoded by the pslA gene which are the determinants of the stability of the biofilm structure. The formation of Pseudomonas aeruginosa biofilm is influenced by several factors, including nutrient composition, environmental conditions (pH and growth temperature), and other inhibitor compounds.(4) Prevalence Pseudomonas aeruginosa mucoid type and non-mucoid type were 3,6 % and 96,4 % respectively.(5) Non-mucoid strains can be involved in the early stages of biofilm development (acute stage of infection), PSL encoded by genes PSL A neutrally charged that supports the structure during the early phase of biofilm development and promoting cell-to-substrate and cell-to-cell interactions binding. Alginateen coded by genes al Dencodes the production of “stress” response polysaccharides associated with chronic infection. Production of alginate which in excess produces a mucoid phenotype and is a significant virulence factor in pulmonary infections. Pseudomonas aeruginosa 5cyctic fibrosis(CF).(3,5)
Pelencoded by genes pelA, is a linear polymer-N-acetylation part ofɑ-1,4-N-acetyl galactosamine which mostly consists of repetition. Dimergalactosamine And N-acety lgalactosamine. Expression of polysaccharide coding loci (pelA) by Pseudomonas aeruginosa is very important for biofilm formation.(5) PelA plays a key role in the extracellular polysaccharide structure of the biofilm matrix to enhance binding to surfaces and other cells, to construct and provide a scaffold to maintain the biofilm structure, and to protect cells from antimicrobial and host defense mechanisms.(6)
A study in Ismailia Mesir reported the frequency of pelA in biofilm-producing strains. Of the 82/95 isolates (86,4 %) the biofilm-producing strains expressed the pelA gene, while 27/31 isolates (87 %) expressed the pelA gene. Strains that express this gene do not produce biofilm – strains that do produce biofilm have the pelA gen.(6)
Biofilm density is the density of the biofilm on the surface. Microtiter plate which is dissolved. Microtiter plates are a quantitative method as a gold standard biofilm test and the most frequently used method. This method has a sensitivity of 97,1 %, specificity of 97,5 %, and accuracy of 97,2 % in detecting biofilm attachment. Its application is quite easy, the price is relatively affordable, and the results can be known quickly. The disadvantage of this method is that the crystal violet material binds to the entire surface of negatively charged molecules and polysaccharides found in the extracellular matrix, including living and dead cells. This crystal violet staining method makes it difficult to determine how many biofilms are dead.(6,7)
Technique Polymerase Chain Reaction (PCR) is a technique for synthesizing and amplifying DNA in vitro. PCR amplification is carried out to identify the presence of genes related to biofilms such as genesis, pelA, and And alg Din clinical isolates of Pseudomonas aeruginosa. The PCR tool has a fairly high sensitivity and specificity, namely a sensitivity of 97,78 % with a specificity of 61,82 %.(7) The limitations of PCR are that it requires special equipment, takes longer, and is not cheap.(8)
Previous studies have focused on the presence or absence of biofilm genes in biofilm-producing bacteria. Research on the relationship between the pelA gene and biofilm density in clinical isolates of Pseudomonas aeruginosa has not been conducted in Indonesia, especially in Surabaya. Based on the description above, researchers are interested in conducting research on the relationship between the pelA gene and biofilm density in clinical isolates of Pseudomonas aeruginosa.
METHOD
Ethical Approval
This research design uses an observational analytical research design using clinical isolates. Pseudomonas aeruginosa at the Clinical Microbiology Unit of Airlangga University Hospital. This study was approved by the Health Research Ethics Committee of Airlangga University Hospital through letter number: 098/KEP/2024.
Sample
Clinical isolates that have been identified pseudomonas aeruginosa were collected and stored in a cryotube containing TSB liquid media and 20 % glycerol, then stored in a freezer with a temperature of -80℃at the Clinical Microbiology Unit of Airlangga University Hospital. The sample collection process was carried out from July 2024 – September 2024. Samples that had been collected during that time were continued to the next stage by examining the biofilm density using the ELISA tool. Reader (BioRad iMark Microplate Reader) and gene detection pelA with PCR (SimpliAmpTMThermal Cycler) were conducted at the Dengue Laboratory of the Institute of Tropical Diseases, Airlangga University.
Preparation of Microbial Samples
Clinical isolate samples of Pseudomonas aeruginosa obtained from the Clinical Microbiology Unit of Airlangga University Hospital were put into Crotube which contains medium Tryptic Soy Broth. (TSB) and 20 % glycerol stored at 37℃ for the next 24 hours stored infreezer-80℃. Bacterial isolates in a medium Soy Broth (TSB) and 20 % glycerol were taken using a sterile loop needle inserted into the media.MacConkey agar for bacterial growth in incubation at a temperature of 37℃for 24 hours. Bacterial isolates grown in the media MacConkey garden subcultured on media MacConkey garden incubated at a temperature of 37℃ for 24 hours.
Biofilm Assay
After subculture for 48 hours, bacterial colonies were taken using a cotton swab and then put into a test tube containing 0,45 % NaCl solution as a standard solution McFarland 0,5 and readings were made on Densi CHEK Plus. The tube containing 1,980 µl Tryptic Soy Broth (TSB) added bacterial suspension Pseudomonas aeruginous as much as 20 µl, then put into each well microtiter plate as much as 200 µl. The PAO1 positive control sample was inserted into the well microtiter plate as much as 200 µl, while the negative control sample contained sterile distilled water of as much as 200 µl. Microtiter plate covered and incubated at 37℃for 24 hours. The well is then washed using Phosphate Buffer Saline (pH 7,2) three times and then dried by turning it over to dry in the open air. The bound bacterial biofilm was well fixed with 150 µl of 96 % methanol for 20 minutes. A microtiter plate was patted at each washing process to remove the solution from the well. Stain the biofilm with 1 % Crystal violet for 5 minutes. The excess Crystal violet dye solution was rinsed gently 3 times with distilled water and plate-dried. Biofilms attached to the walls and bottom of the tube that have been stained with crystal violet will be dissolved with 150 µl of 96 % ethanol in each well. Close the microtiter plate slowly, and the preparation is left at room temperature for 5 minutes. The thickness of the biofilm bound to the bottom of the well microtiter plate was read by an ELISA reader with the value Optical Density read at λ 595 nm.
Data Processing
The strength of the test bacterial biofilm determines the OD value. Isolate compared to the value Optical Density Cut-off(O.D.cut). The OD value is obtained from the average value, and the OD value isolates minus the OD value cut (OD = average value of ODisolate – ODcut). ODcut defined as three standard deviations above the mean OD of the control (culture medium) (Ruchi, Sujata, and Anuradha, 2015). Formula for calculating ODcut as follows: ODcut = x̄ ODControl+ (3 x SDControl) Information:
ODcut = Optical Density cut-off
x OD Control = Optical Density control (Average optical density value of the Wells microplate without adding inoculum)
SD Control = Standard Deviation from the well without adding inoculum
|
Table 1. Interpretation of the Strength of Biofilm-Forming Bacteria (July 07, 2024, Surabaya) |
|
|
Average OD value |
Biofilm Producing Power |
|
O.D.isolate≤ ODcut |
Biofilm not produced (0) Weak |
|
O.D.cut<O.D.isolate≤ 2x ODcut |
Biofilm production (+) Moderate |
|
2x ODcut<O.D.isolate≤ 4 x ODcut |
Biofilm production (++) |
|
4x ODcut<ODisolate |
Strong biofilm production (+++) |
|
Note: ODcut = Optical Density cut, OD isolate = Optical Density isolate |
|
pelA Gene Detection
DNA Isolation
Bacterial colonies on media Mac Conkey Agar (MCA) were taken as much as 3-5 colonies and the colonies were put into a microcentrifuge tube containing 180µl. Buffer Testand 20µlPK solution then mix for 20 seconds. The sample was incubated at a temperature of 56℃ for 30 minutes. The sample was added 200µl LysisB buffer and mixed well. The sample was added with 200µl of absolute ethanol and mixed well.
DNA Purification
Transfer all sample mixtures ≤750µl inside Column. The sample mixture was centrifuged at 12 000 rpm for 1 minute. Add 500µl of sample Buffer into the theSpin Column. The sample mixture was centrifuged at 12 000 rpm for 1 minute. Add 500µl sample Buffer into the theSpin Column. The sample mixture was centrifuged at 12 000 rpm for 1 minute. Repeat step 3 1x. The spin column was centrifuged at 12,000 rpm for 2 minutes. The centrifuged sample was added 50-100µl Elution Buffer into the spin Column and incubated at room temperature for 1 minute and then centrifuged at 12 000 rpm for 1 minute.
The liquid in the tube containing DNA, the extracted DNA can be stored at room temperature. -20 ℃ and used for PCR examination samples.
pelA Gene Detection
1. Settings PCR machine: Pre-Denaturation 95℃for 5 minutes (1 cycle), Denaturation 95℃ for 30 seconds, Annealing 59℃for 30 seconds, Extension 72℃for 1 minute, Final Extension72℃for 5 minutes (1 cycle), Cooling20℃for 30 seconds (1 cycle).
2. Preparing PCR mix:
Remove the Master mix (NEXproTM e PCR 2X Master Mix) from the freezer to be vortexed, then the master mix is spun down slowly. The composition of the material checked in PCR: Master mix (NEXproTM e PCR 2X Master Mix) as much as 12,5 µl, 1 µl of forward primer pelA (Macrogen), 1 µl of reverse primer pelA (Macrogen), 7,5 µl of ddH2O. The total final volume is 22 µl/sample tube. The sample is PCRed in a 1,5 ml Eppendorf tube then vortexed, spun down, and pipetted into each tube as much as 22 µl/tube. The DNA template (sample) is pipetted as much as 3 µl which has been filled with 22 µl of PCR mix so that the final volume of each tube is 25 µl.
3. Process running done by entering PCR.
4. Program running PCR is selected according to primary settings select “RUN” then “OK”, and wait until the PCR process is complete. (9)
Gene Detection pelA using the primary sequence:
Table 2. Gene Primer pelA with PCR (July 07, 2024, Surabaya) |
|||
Gene |
Primary |
Primary Sequence (5’-3’) |
Amplicon Size(bp) |
pelA |
F |
CCTTCAGCCATCCGTTCTTCT |
118bp |
|
|
R |
TCGCGTACGAAGTCGACCTT |
|
Source: (Colvinet al.,2012) |
|||
The PCR results were then electrophoresed with 2 % agarose gel electrophoresis the sample was mixed with as much as 2 µlloading dye. Agarose gelprinted and after solidifying, inserted into the electrophoresis container then inserted 6 µl of each sample and NEXmark 100bp Ladderas much as 6 µl. Agarose gel was then electrophoresed at 100 volts for 30 minutes and then appeared on a UV transilluminator. PCR result positive gene pelA and positive control (Pseudomonas aeruginosaPAO1) is indicated by the appearance of DNA fragments of 118 bp each. Negative control using RNA seawater free. PCR amplification using primers pelA has a target amplification region of 118 bp.(10)
Statistical Methods
Statistical data analysis was carried out using correlation analysis using the test Fisher’s Exact test to analyze the relationship between genes pelA and biofilm density in clinical isolates of Pseudomonas aeruginosa in the SPSS version 25 program. To see the significance, the statistical test test Fisher’s Exact with values-Value=0,05. Analysis was also carried out using analysis contingency coefficient to find out how strong the relationship is between genes pelA with biofilm density in clinical isolates of Pseudomonas aeruginosa.
RESULTS
Distribution in Clinical Isolates Pseudomonas aeruginosa
Clinical isolate distribution data Pseudomonas aeruginosa obtained from the logbook. Examination of specimen culture at the Clinical Microbiology Unit of Airlangga University Hospital during The research period was July – September 2024. Clinical isolates Pseudomonas namely Pseudomonas aeruginosa with percent probability≥ 93 % examined with toolsVitek 2 Compact (bioMerieux). This research was conducted on 33 isolates of clinical Pseudomonas obtained from various types of specimens of which 22 (66,7 %) were from sputum, 3 (9,1 %) were from pus, 2 (6,1 %) were from blood, 6 (18,2 %) were from urine.
|
Table 3. Distribution of Clinical Isolates Pseudomonas aeruginosa (September 09, 2024, Surabaya) |
||
|
Spesimen |
Frekuensi |
Persentase |
|
Sputum |
22 |
66,7 % |
|
Pus |
3 |
9,1 % |
|
Darah |
2 |
6,1 % |
|
Urine |
6 |
18,2 % |
|
Total |
33 |
100 % |
Colony Morphology in Clinical Isolates of Pseudomonas aeruginosa
Colony Morphology in Clinical Isolates Pseudomonas aeruginosa Non-mucoid strains, such as pseudomonas aeruginosa, have flat and uniform colonies, while the mucoid strain, pseudomonas aeruginosa, produces alginate. Excessive (mucoid) has a slimy appearance and a thick and viscous structure. The results Analysis of 33 clinical isolates, such as pseudomonas, shows the number of strains. Non-mucoid as many as 32 (97,0 %) and mucoid strains Pseudomonas aeruginosa As many as 1 (3,0 %) in sample code S.21.
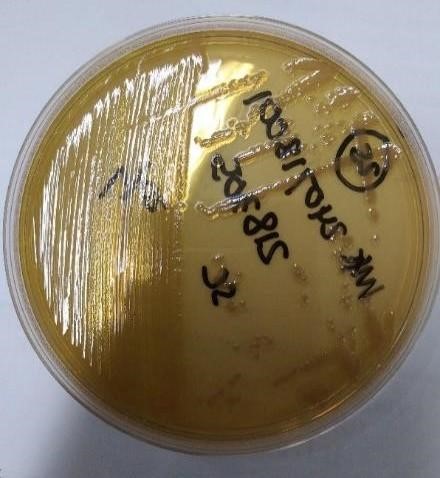
Figure 1. Non-Mucoid Morphology. (Airlangga University Hospital, Surabaya, 2024)
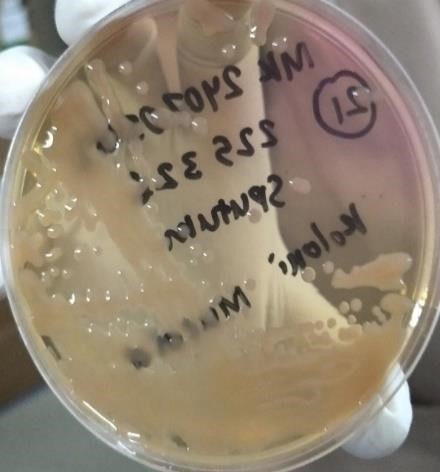
Figure 2. Mucoid Morphology (Airlangga University Hospital, Surabaya,
2024)
Gene Detection Results pelAIn Clinical Isolates Pseudomonas aeruginosa
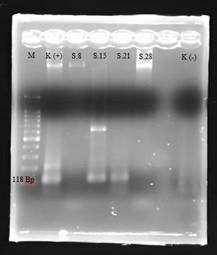
Figure 3. Agarose Gel Electrophoresis (2 %) of the PCR Product pelA Gene (118 bp) (Airlangga University Hospital, Surabaya, 2024)
|
Table 4. Number of Gene Visualization Results pelA Clinical Isolate Pseudomonas aeruginosa (September 09, 2024, Surabaya) |
||
|
Number of clinical isolates Pseudomonas aeruginosa |
Gene pelA (+) |
Gene pelA (-) |
|
33 |
31 (93,9 %) |
2 (6,1 %) |
|
Table 5. Gene Results pelA And Biofilm Density (September 09, 2024, Surabaya) |
||
|
Biofilm Density |
Gene pelA(+) |
GenepelA(-) |
|
Not Produced |
2 (50 %) |
2 (50 %) |
|
Weak |
10 (100 %) |
0 (0 %) |
|
Currently |
13 (100 %) |
0 (0 %) |
|
Strong |
6 (100 %) |
0 (0 %) |
Biofilm Density Examination Results on Clinical Isolates Pseudomonas aeruginosa
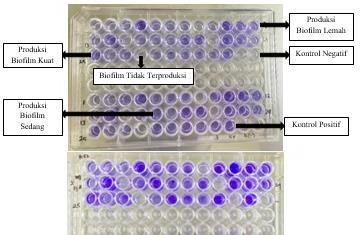
Figure 4. Biofilm Density Examination Method Microtiter Plate (Airlangga University Hospital, Surabaya 2024)
|
Table 6. Biofilm Density of Clinical IsolatesPseudomonas aeruginosa (September 09, 2024, Surabaya) |
||
|
Biofilm Density |
Number of Isolates |
Percentage of Isolates |
|
Not Produced |
4 |
12,1 % |
|
Weak |
10 |
30,3 % |
|
Currently |
13 |
39,4 % |
|
Strong |
6 |
18,2 % |
|
Total |
33 |
100 % |
Relationship Between Genes pelA And Biofilm Density In Clinical Isolates Pseudomonas aeruginosa
Then it can be concluded that there is a relationship between genes pelA and biofilm density in clinical isolates of Pseudomonas aeruginosa. The analysis contingency coefficient obtained a value of 0,565 with a value of approximate sig. p-Value=0,001 means that the strength of the relationship between genes pelA and biofilm density in clinical isolates Pseudomonas aeruginosais moderate.
|
Table 7. Relationship Between Genes pelA and Biofilm Density (September 09, 2024, Surabaya) |
||||
|
Biofilm Density |
Gene pelA (+) |
GenepelA(-) |
p-Value |
Test Type |
|
Not Produced |
2 (50 %) |
2 (50 %) |
0,011 |
Fisher Exact |
|
Weak |
10 (100 %) |
0 (0 %) |
||
|
Currently |
13 (100 %) |
0 (0 %) |
||
|
Strong |
6 (100 %) |
0 (0 %) |
||
|
Note: Statistical analysis with tests Fisher’s Exact Test gives value-Value=0,011 |
||||
Relationship between Genes pelA and Biofilm Density in Clinical Isolates Pseudomonas aeruginosa in Non-Mucoid Strains
It can be concluded that there is a relationship between the pelA gene and biofilm density in clinical isolates of Pseudomonas aeruginosa in non-mucoid strains, whereas in mucoid strain could not be statistically analyzed because only 1 was found Mucoid strain. The contingency coefficient analysis obtained a value of 0,564 with a value of approximate sig. P-Value = 0,002 means that the strength of the relationship between genes pelA and biofilm density in clinical isolates of Pseudomonas aeruginosa in non-strains mucoid is moderate.
|
Table 8. Relationship between Genes pelA and Biofilm Density in Non-Mucoid Strains (September 09, 2024, Surabaya) |
|||||
|
|
Density Biofilm |
GenepelA (+) |
GenepelA (-) |
p-Value |
Type Test |
|
Non-Mucoid |
No Produced |
2 (50 %) |
2 (50 %) |
0,012 |
Fisher’S Exact |
|
|
Weak |
10 (100 %) |
0 (0 %) |
||
|
|
Currently |
13 (100 %) |
0 (0 %) |
||
|
|
Strong |
6 (100 %) |
0 (0 %) |
||
|
Note: Statistical analysis using Fisher’s Exact Test gave a p-value = 0,012. |
|||||
DISCUSSION
This study was conducted on 33 clinical isolates. Pseudomonas aeruginosa, which obtained from various types of specimens including 22 (66,7 %) from sputum, 3 (9,1 %) from pus, 2 (6,1 %) from blood, 6 (18,2 %) from urine. Clinical isolates that have been identified as Pseudomonas aeruginosa own percent probability≥ 93 % examined with toolsVitek 2 Compact (bio Merieux). Isolation of bacteria is done by carrying out inoculation on Media Mac Conkey Agar(MCA), which was then incubated for 24 hours at a temperature of 37℃. The process of bacterial growth and isolation process to obtain clinical isolates Pseudomonas aeruginosa which has 32 (97,0 %) non-mucoid strains and mucoid strains as many as 1 (3,0 %) with sample code S.21, to see the morphology non-mucoid and mucoid strains Pseudomonas aeruginosa viewed macroscopically. Isolate Pseudomonas aeruginos which is a non-mucoid strain has colonies that are flat, and uniform, and produce flat, dense biofilms, while mucoid strains have a slimy appearance, has a thick and viscous structure. Pseudomonas strains aeruginosa produces excess alginate (mucoid) and has a mucus-like appearance and Alginate plays a role in the formation of thick and three-dimensional biofilms.
The pelA gene encodes pel for the production of polysaccharides that are important in biofilm formation. Pel and Psl are the main structural polysaccharides of biofilms. Pseudomonas aeruginosa non-mucoid and mucoid.(9) Components non-mucoid Pseudomonas aeruginosa biofilm supporters other than exopolysaccharides, eDNA which is abundant can increase cell surface attachment and maintain In 3D architecture, eDNA can be used as a supporting component of biofilms to provide nutrients for bacteria in biofilms.(10)
Non-mucoid strains in Biofilm production are characterized by high levels of c-di-GMP. Pseudomonas aeruginosa’s high levels of biofilm formation correlate with high levels of c-di-GMP. The secondary messenger c-di-GMP promotes the production of biofilm components such as PSL and peel. Mediation by c-di-GMP regulates all extracellular matrix components of biofilms such as exopolysaccharides and eDNA. Strains that fail to produce pel, do not have a functional PSL operon. The levels of c-di-GMP which is low correlated with low biofilm counts.(11)
Alginate, PSL, and Andmopare three exopolysaccharides that are responsible for the main components in the biofilm matrix especially those related to protecting bacterial cells from antibiotics and the human immune system. Pseudomonas aeruginosa own One of the virulence factor genes involved in biofilm formation is the gene pelA. Gen pelA has a function that plays an important role in the formation of structures of carbohydrate-rich biofilm matrix, maturation, and maintaining biofilm, genes pelA can mediate cell-to-cell interactions through its positive cross-linking charge eDNA is negatively charged through ionic interactions. Cationic charges play an important role in its ability to interact with other major biofilm matrix components.(12)
Results of gene detection pelA The results obtained were 31 (93,9 %) isolates. Positive gene pelA and as many as 2 (6,1 %) isolates were negative for the gene pelA. Based on the results of Biofilm density examination and gene detection pelA against 33 clinical isolates Pseudomonas aeruginosa got positive results with genepelAconsists of 2 (50 %) samples that did not produce biofilm, as many as 10 (100 %) samples produced biofilm weak, as many as 13 (100 %) samples produced moderate biofilm, and as many as 6 (100 %) samples produced strong biofilm. Negative gene results in many as 2 (50 %) samples did not produce biofilm. Abdel Raheem’s research results al., (2020) stated that of the 24 isolates (24 %) that expressed the gene pelA, as many as 18 isolates (66,7 %) from biofilm-producing strains while 6 isolates (8,2 %) from non-biofilm-producing-strains-Producing-biofilms.(13)
Informed that 80 % (100/126) of clinical isolates were positive biofilm makers using the positive pelA gene consisting of 22 (18 %) were strong biofilm makers, 43 (34 %) were medium biofilm makers, 35 (28 %) were weak biofilm makers producers & 31 (20 %) were non-biofilm producers.(14) Clinical isolates are unable to form biofilms despite carrying the genepelA because there are other genes responsible for biofilm formation.(15) Many studies have investigated other types of biofilm genes. The results of the AL-Sabawi.(16) found that the gene pelF is responsible for the formation of biofilm.(17)
Pseudomonas aeruginosa is associated with several infections in humans, especially those related to health services, one of the biggest challenges in treating infection Pseudomonas aeruginosa is associated with biofilms.(18) The formation of biofilms in this study was carried out by cultivating biofilms on a microtiter plate and each well was inoculated with a clinical isolate that has been formed as a suspension followed by measurement of biofilm density as many as 3x repetitions using the tool BioRad iMark Microplate Readeron λ595 nm was examined at the Research Laboratory Installation of Airlangga University Hospital. Based on the research that has been carried out, the total biomass value can be calculated with crystal violet staining. The mechanism of this method is the presence of molecules negatively charged surfaces of both living and dead cells, and exopolysaccharides The biofilm that is formed is bound by crystal violet dye so that it is produced Absorbance value.(18,19)
This study was conducted on 33 clinical isolates. Pseudomonas aeruginosa. This study obtained results that 4 (12,1 %) clinical isolates did not produce biofilm, 10 (30,3 %) clinical isolates had weak biofilm production, 13 (39,4 %) clinical isolates had weak biofilm production moderate biofilm, 6 (18,2 %) clinical isolates had strong biofilm production. Biofilm examination is done by the method microtiter plate. Previous studies have reported a high level of different biofilm production by isolatesPseudomonas aeruginosa in Egypt regarding biofilm formation was detected in 32/35 (91,4 %) isolates Pseudomonas aeruginosa namely 25,7 %, 40 %, 25,7 %, and 8,6 % of isolates were biofilm producers respectively strong, medium, weak, and non-biofilm producers.(20) This research is by research by Harika et who reported that 78,2 % (72/92) of clinical isolates of Pseudomonas aeruginous a biofilm producer positive, 69,5 % (64/92) produced strong biofilm, 8,7 % (8/92) produced biofilm moderate, and 21,7 % (20/92) produced weak biofilms.(21,22)
The results of this study found that the gene pelA has a relationship with Density Biofilms in Clinical Isolates Pseudomonas aeruginosa which has been subjected to statistical testing using test Fisher’s Exact test the SPSS version 25 program. The results obtained for values-Value=0,011 then it can be concluded that there is a relationship between Gene detection pelA and biofilm density in clinical isolates Pseudomonas aeruginosa. The contingency coefficient analysis obtained a value of 0,565 with a value of approximately sig. p-Value=0,001 then it can be concluded that there is a strong relationship Moderate. Clinical isolate Pseudomonas aeruginosa has high virulence genes and the ability to form biofilms, most studies show a correlation significant relationship between biofilm formation and current virulence genes.(23,24,25)
The results of this study found that the gene pelA has a relationship with Density Biofilms in Clinical Isolates of Pseudomonas aeruginosa in non-mucoid strains, whereas the mucoid strain could not be analyzed because only 1 mucoid strain was found. The contingency coefficient analysis obtained a value of 0,564 with a value of approximately sig. p –Value=0,002, it can be concluded that there is a strong relationship medium. The results of Elmaraghy’s found that the strain producing certain biofilms is specifically associated with certain virulence genes. Nineteen strains of 23 (82,6 %) expressed the gene pelA with sig. P-value statistically (P≤0,05).(26)
Research limitations
Not testing Pel levels produced by mucoid and non-mucoid strains. Did not measure the concentration of c-di-GMP in biofilm formation in non-mucoid strains. Not detecting other genes that play a role in biofilm formation in Pseudomonas aeruginosa isolates, namely detecting the pslA gene and the algD gene.
CONCLUSION
Based on the research results obtained in this study, the following conclusions can be outlined:
The pelA gene is associated with biofilm density in clinical isolates of Pseudomonas aeruginosa with moderate strength of association. The pelA gene is associated with biofilm density in clinical isolates of Pseudomonas aeruginosa in non-mucoid strains with moderate strength of association.
BIBLIOGRAPHIC REFERENCES
1. Abdelraheem, W.M.et al.(2020) ‘Detection of biofilm formation and assessment of biofilm genes expression in different Pseudomonas aeruginosa clinical isolates’, Meta Gene, 23. doi: 10.1016/j.mgene.2020.100646.
2. Al-Draghi, WAH and Saeed, RAAK (2020) ‘Synergistic effect of amikacin and ciprofloxacin on PelA and AlgD genes in pseudomonas aeruginosa’, Indian Journal of Forensic Medicine and Toxicology, 14(4), pp. 1608–1614. doi: 10.37506/ijfmt.v14i4.11772.
3. Al-Sabawi, BAM (2017) gene expression variation of some virulence factors of pseudomonas aeruginosa isolated from several pathogenic and environmental cases. University of Al-Anbar, Iraq.
4. Al-Sheikhly, MAARH, Musleh, LN and Al-Mathkhury, HJF (2019) ‘Assessment of pelA-carried pseudomonas aeruginosa isolates in respect to biofilm formation’, Iraqi Journal of Science, 60(6), pp. 1180–1187. doi: 10.24996/ijs.2019.60.6.1.
5. AL-Sheikhly, MARH, Musleh, LN and Al-Mathkhury, HJF (2020) ‘Gene expression of pelA and pslA in Pseudomonas aeruginosa under gentamicin stress’, Iraqi Journal of Science, 61(2), pp. 295–305. doi: 10.24996/ijs.2020.61.2.6.
6. Balducci, E.et al.(2023) ‘Polysaccharides’ Structures and Functions in Biofilms Architecture of Antimicrobial-Resistant (AMR) Pathogens’, International Journal of Molecular Sciences, 24(4). doi: 10.3390/ijms24044030.
7. Cho, HHet al.(2018) ‘Association between biofilm formation and antimicrobial resistance in carbapenem-resistant Pseudomonas aeruginosa’, Annals of Clinical and Laboratory Science, 48(3), pp. 363–368.
8. Colvin, K.M.et al.(2012) ‘The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix’, Environmental Microbiology, 14(8), pp. 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x.
9. Colvin, K.M.et al.(2013) ‘PelA deacetylase activity is required for pel polysaccharide synthesis in pseudomonas aeruginosa’, Journal of Bacteriology, 195(10), pp. 2329–2339. doi: 10.1128/JB.02150-12.
10. Costa, ACBP, et al. (2013) ‘Methods for obtaining reliable and reproducible results in studies of Candida biofilms formed in vitro’, Mycoses, 56, pp. 614–622. doi: 10.1111/myc.12092.
11. Deligianni, E.et al.(2010) ‘Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro’, BMC Microbiology, 10, pp. 1–13. doi: 10.1186/1471-2180-10-38.
12. Elmaraghy, N., Abbadi, S. and Elhadidi, G. (2019) ‘Virulence Genes in Pseudomonas Aeruginosa Strains Isolated at Suez Canal University Hospitals concerning the Site of Infection and Antimicrobial Resistance’, Int J Clin Microbiol Biochem Technol, 2, pp. 008–019. doi: 10.29328/journal.ijcmbt.1001006.
13. Farhan, REet al. (2023) ‘Molecular detection of different virulence factors genes harbor, pelA, exoS, toxA and algD among biofilm-forming clinical isolates of Pseudomonas aeruginosa’, Cellular and Molecular Biology, 69(5), pp. 32– 39. doi: 10.14715/cmb/2023.69.5.6.
14. Ghafoor, A., Hay, I.D. and Rehm, B.H.A. (2011) ‘Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture’, Applied and Environmental Microbiology, 77(15), pp. 5238–5246. doi: 10.1128/ AEM.00637-11.
15. Ghazalibina, M.et al.(2019) ‘Study of virulence genes and association with biofilm formation in Pseudomonas aeruginosa isolated from clinical samples of Iranian patients’, Pre-Proof (Gene Reports), p. 100471. doi: 10.1016/j.genrep.2019.100471.
16. Harika, K.et al.(2020) ‘Detection of Biofilm Production and Its Impact on Antibiotics Resistance Profile of Bacterial Isolates from Chronic Wound Infections’, Journal of Global Infectious Diseases, (November 2019), pp. 129–134. doi: 10.4103/jgid.jgid.
17. Ismaun, Muzuni, and Hikmah, N. (2021) ‘Molecular Detection of Escherichia coli Bacteria As a Cause of Diarrhea Disease Using PCR Technique’, Biome, 6(2), pp. 1–9.
18. Jennings, L.K.et al.(2015) ‘Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix’, Proceedings of the National Academy of Sciences of the United States of America, 112(36), pp. 11353–11358. doi: 10.1073/pnas.1503058112.
19. Malhotra, S.et al.(2019) ‘Cystic Fibrosis and Pseudomonas aeruginosa: the Host- Microbe Interface’,Clinical Microbiology Reviews, 32(3), pp. 1–46. doi: https://doi.org/10.1128/cmr.00138-18.
20. Pratiwi, SUTet al.(2015) ‘Effect of Cinnamomum burmannii Nees ex Bl. and Massoia aromatica Becc. Essential oils on planktonic growth and biofilm formation of Pseudomonas aeruginosa and Staphylococcus aureus In Vitro’, International Journal of Applied Research in Natural Products, 8(2), pp. 1–13.
21. Ramli, S. (2020) Relationship Between Biofilm Density and pslA Gene in Clinical Isolates Pseudomonas aeruginosa at Dr. Soetomo Hospital, Surabaya. Airlangga University.
22. You, FFet al.(2022) ‘Pathogenesis of the Pseudomonas aeruginosa Biofilm : A Review’, Pathogens, 11(300)
23. Santo Guanoluisa JM, Ron Mora Álvaro S. Bacterial resistance of microbial agents causing urinary tract infections in pregnant women. Salud, Ciencia y Tecnología. 2024;4:728.
24. Ali Naser A, Jassim AlSultany S. Multidrug Resistant Bacteria Isolated from Urinary Tract Infections in Pregnancy Association with C -Reactive Protein. Salud, Ciencia y Tecnología. 2024;4:1294.
25. Wei, Q. and Ma, L.Z. (2013) ‘Biofilm matrix and its regulation in Pseudomonas aeruginosa’,Int. J. Mol. Sci, 14(10), pp. 20983–21005. doi: 10.3390/ijms141020983.
26. Zhao, A., Sun, J. and Liu, Y. (2023) ‘Understanding bacterial biofilms: From definition to treatment strategies’, Frontiers in Cellular and Infection Microbiology, 13, pp. 1–23. doi: 10.3389/fcimb.2023.1137947.
FINANCING
The authors did not receive financing for the development of this research.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORSHIP CONTRIBUTION
Conceptualization: Nanda Yaultsa.
Data curation: Nanda Yaultsa, Puspa Wardhani, Aryati.
Formal analysis: Nanda Yaultsa.
Research: Nanda Yaultsa.
Methodology: Nanda Yaultsa, Puspa Wardhani, Aryati.
Project management: Nanda Yaultsa, Puspa Wardhani, Aryati.
Resources: Nanda Yaultsa, Puspa Wardhani, Aryati.
Software: Nanda Yaultsa.
Supervision: Puspa Wardhani, Aryati.
Validation: Puspa Wardhani, Aryati.
Display: Puspa Wardhani, Aryati.
Drafting - original draft: Nanda Yaultsa.
Writing - proofreading and editing: Nanda Yaultsa, Puspa Wardhani.